Fig. 6.
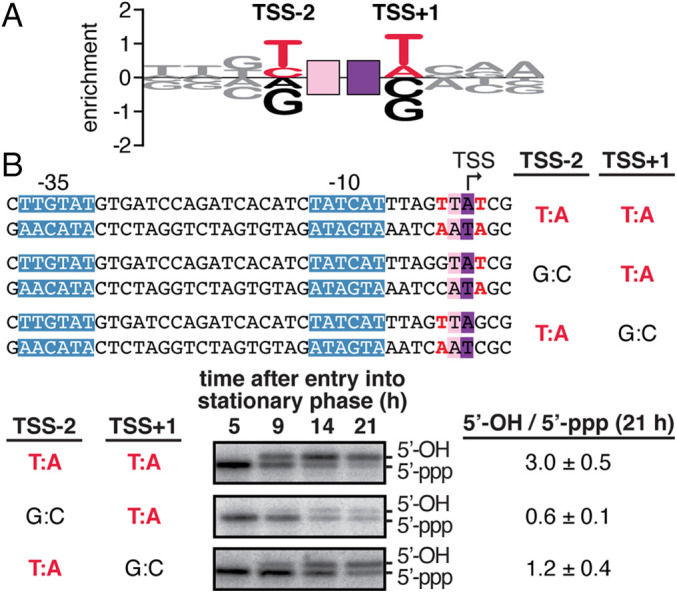
Promoter-sequence dependence of primer-dependent initiation: chromosomal promoters. (A) Sequence logo for primer-dependent initiation at TSS positions 7, 8, and 9 (corresponding to primer binding sites 6 to 7, 7 to 8, and 8 to 9, respectively) in stationary-phase E. coli cells for 93 natural, chromosomally encoded promoters that use UpA as a primer. The height of each base X at each position Y represents the relative log2 enrichment (averaged across replicates) of the percent 5′-OH RNAs expressed from promoter sequences containing nontemplate-strand base X at position Y. Red, consensus nucleotides; black, nonconsensus nucleotides. Other symbols and colors as in Fig. 1. (B) Promoter-sequence dependence of primer-dependent initiation at the E. coli bhsA promoter. (Top) Sequences of DNA templates containing wild-type and mutant derivatives of bhsA promoter. (Bottom) Primer extension analysis of 5′-end lengths of bhsA RNAs. In primer-dependent initiation with a dinucleotide primer, the RNA product acquires one additional nucleotide at the RNA 5′-end (Fig. 1). Gel shows radiolabeled cDNA products derived from primer-independent initiation (5′-ppp) and primer-dependent initiation (5′-OH) in stationary-phase E. coli cells. (Bottom Right) Ratios of primer-dependent initiation versus primer-independent initiation (mean ± SD, n = 4).
