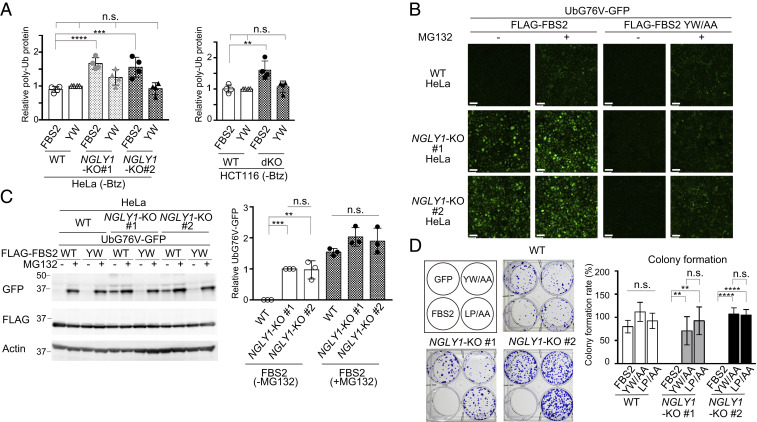
Fig. 3.
FBS2 overexpression induces proteasomal dysfunction in NGLY1-KO cells. (A) Accumulation of ubiquitinated proteins in NGLY1-KO HeLa (#1, exon 6 mutant; #2, exon 11 mutant) and HCT116 (FBS2;NGLY1-dKO) cells by FBS2 overexpression. Cells were treated with or without 20 nM bortezomib (Btz) for 5 h prior to harvest. The relative levels of ubiquitinated proteins in cell lysates in SI Appendix, Fig. S3 A and B were quantified. The intensity of the lane with wild-type (WT) cells overexpressing the YW/AA mutant was defined as 1. Error bars show means ± SD of four biological replicates. (B) Low-magnification fluorescence micrographs of WT and NGLY1-KO HeLa cells coexpressing UbG76V-GFP and FLAG-FBS2 (WT or YW/AA mutant) treated with or without 50 μM MG132. (Scale bars, 100 µm.) (C) Detection of UbG76V-GFP accumulation by immunoblotting. Accumulation of UbG76V-GFP in FBS2-overexpressing NGLY1-KO cells. (Left) Cell lysates in (B) were analyzed with anti-GFP antibody. (Right) Relative levels of UbG76V-GFP in FBS2-overexpressing WT and NGLY1-KO cells were quantified; the intensity of the band of FBS2-overexpressing NGLY1-KO #1 cells without MG132 treatment was defined as 1. Error bars show means ± SD of three biological replicates. (D) Colony formation assays in WT and NGLY1-KO HeLa cells. (Left) Colony formation assay in WT and NGLY1-KO HeLa cells (#1, exon 6 mutant; #2, exon 11 mutant) overexpressing the indicated proteins. Recombinant retrovirus for the expression of the indicated proteins, produced by pMXs-puro, was used to infect WT and NGLY1-KO HeLa cells. After infection for 36 h, 500 cells were plated onto 6-well dishes in the presence of 1 μg/mL puromycin. After culture for 11 to 14 d, the colonies were stained with crystal violet and counted. (Right) Colony formation rates were calculated relative to the number of colonies formed by GFP-overexpressing cells. Cells were counted in each of four replicates. Statistical significance was determined by one-way ANOVA with Dunnett’s posttest (A) or one-way ANOVA with Tukey’s posttest (C and D). ****P < 0.0001; ***P < 0.0002; **P < 0.002; n.s., not significant.