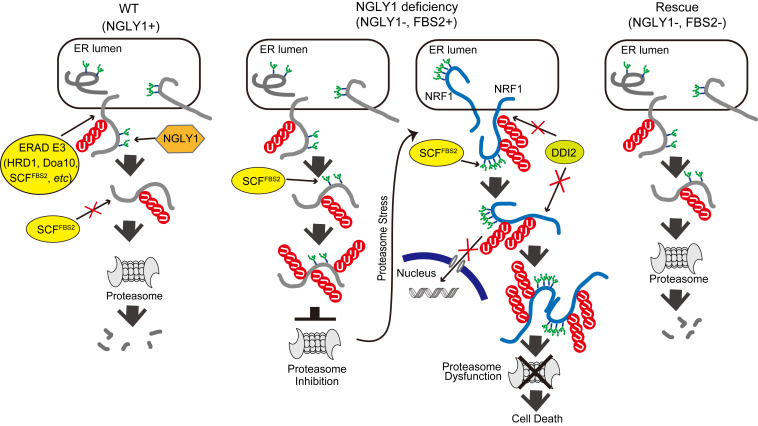
Fig. 6.
Schematic model of the molecular mechanism of induction of cell death by FBS2 in NGLY1-KO cells. (Left) In wild-type (WT) cells expressing NGLY1 with or without FBS2, glycoprotein ERAD substrates are ubiquitinated by multiple ERAD-E3s, such as HRD1, Doa10, or SCFFBS2, upon retrotranslocation to the cytosol. Most ubiquitinated glycoproteins are deglycosylated by NGLY1 prior to proteasomal degradation. (Middle) In NGLY1-KO cells expressing FBS2 (NGLY1 deficiency), glycoproteins that accumulate are ubiquitinated by SCFFBS2. Glycoproteins are constitutively ubiquitinated by SCFFBS2, which overloads the proteasome, thereby impairing it. The cells then sense proteasome stress, and transcriptional activation of proteasome subunits is induced by active NRF1. FBS2 expression causes inactivation of NRF1, which in turn suppresses NRF1-triggered expression of proteasome subunits. As with the ubiquitinated glycoprotein substrates, accumulation of NRF1 itself ubiquitinated by SCFFBS2 causes proteasome and ultimately induces cell death. (Right) In cells lacking both NGLY1 and FBS2, most ERAD substrates are degraded normally, and abnormally processed NRF1 moves to the nucleus without accumulating in the cytosol.