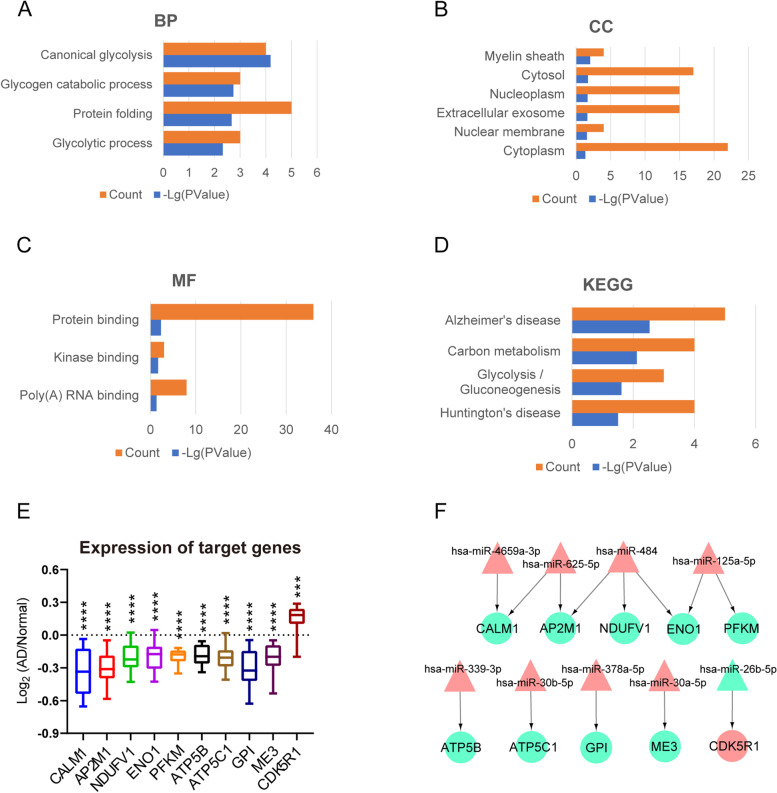
Fig. 5.
Significant biological process (BP) (A), cellular component (CC) (B), and molecular function (MF) (C) terms enriched in target genes of 55 common differentially expressed microRNAs (CDEmiRNAs). KEGG pathway analysis showed the target genes were mainly enriched in Alzheimer's disease, carbon metabolism, glycolysis/gluconeogenesis, and Huntington's disease (D). 10 target genes were found in the KEGG pathway analysis, including CALM1, AP2M1, NDUFV1, ENO1, PFKM, ATP5B, ATP5C1, GPI, ME3, and CDK5R1 (E). CDK5R1 was an increased gene, the other 9 target genes were all decreased its expression in AD samples, *** p < 0.001, **** p < 0.0001. An miRNA–mRNA network of target genes discovered that 9 CDEmiRNAs were involved in the regulation of these 10 target genes (F). The nine downregulated target genes were regulated by hsa-miR-4659a-3p, hsa-miR-625-5p, hsa-miR-484, hsa-miR-125a-5p, hsa-miR-339-3p, hsa-miR-30b-5p, hsa-miR-378a-5p, and hsa-miR-30a-5p, and the increased CDK5R1 was regulated by hsa-miR-26b-5p, red labels represented upregulated miRNAs and target genes, and green labels represented downregulated miRNAs and target genes