Figure 2. Adenosine-to-Inosine RNA editing and current protein-based analytical platforms.
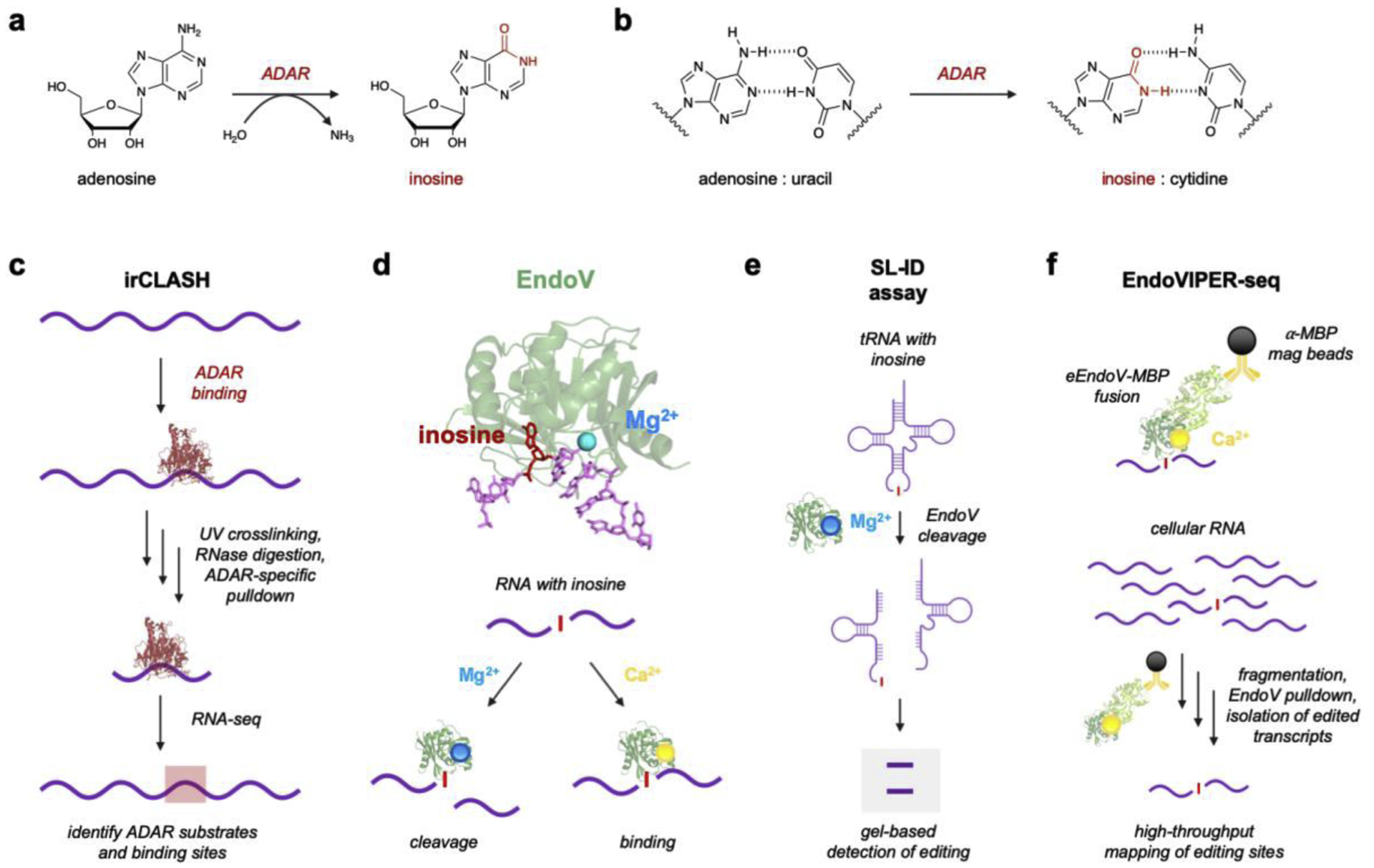
a) A-to-I RNA editing is catalyzed by ADARs via a deamination reaction. b) Resulting inosines base pair with cytidine rather than uracil. c) irCLASH enables large-scale identification of RNA binding sites for different ADAR enzymes. RNA-protein complexes are crosslinked and unbound RNA is digested. Immunoprecipitation (IP) of ADAR-RNA complexes enables sequencing of bound RNA regions. d) Endonuclease V (EndoV) is an inosine-specific ribonuclease. The crystal structure of EndoV (green) from T. maritima (PDB 2W35) is shown complexed with both Mg2+ (blue) and a ssDNA (purple) containing inosine (red). EndoV activity can be modulated by supplementing the enzyme with either Mg2+ for cleavage or Ca2+ for RNA binding. Figure reproduced with permission.53 d) EndoV’s natural cleavage ability with Mg2+ can be used to detect inosine in tRNAs using splinted ligation-based inosine detection (SL-ID). f) EndoV can also be supplemented with Ca2+ and used to bind A-to-I edited transcripts, which was developed into EndoVIPER-seq (EndoV immunoprecipitation enrichment sequencing). Figure reproduced with permission.53
