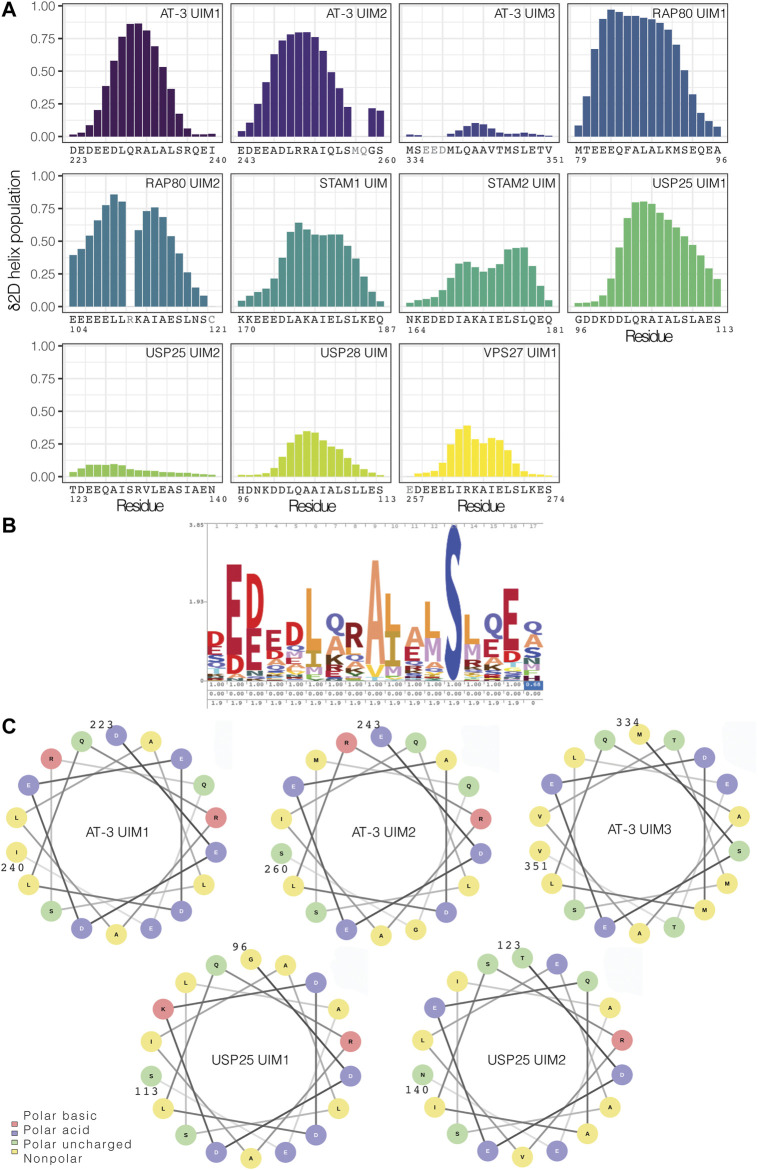
FIGURE 7.
UIMs in the free state can vary from highly helical conformations to disordered counterparts. (A) Helical content for the UIMs predicted from chemical shifts by δ2D. We used nine sets of released chemical shifts of UIMs in the free state in solution from the BMRB database, including AT-3 UIM3, STAM1 UIM, STAM2 UIM, USP28 UIM, USP25 UIM1, and UIM2, and RAP80 UIM1 and UIM2. We also used a set of released chemical shifts of VPS27 UIM1 in fusion with the ubiquitin. In addition, we used the NMR chemical shifts that we recorded for AT-3182-291 UIM1 and UIM2. We highlighted in gray the residues for which there are not enough chemical shifts to run the prediction with δ2D. UIMs have a wide range of predicted helical content. (B) Consensus sequence for UIMs in the PFAM database. (C) Helical wheel representation of AT-3 UIM1, UIM2, and UIM3, and of USP25 UIM1 and UIM2.