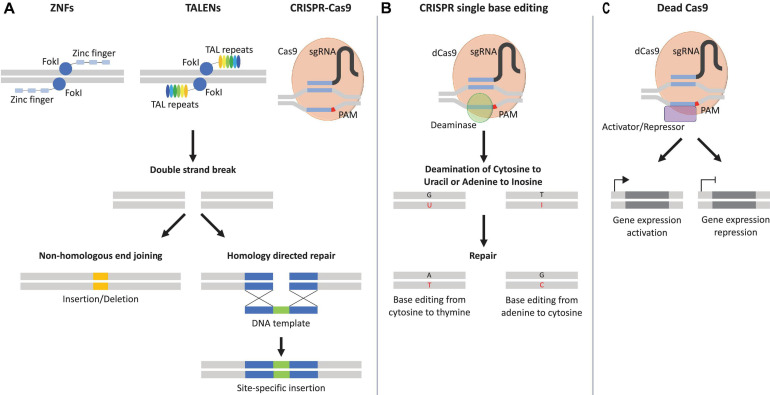
FIGURE 2.
Genome editing tools. (A) Zinc finger nucleases (ZNFs) and transcription activator-like effector nucleases (TALENs) recognize specific genomic sites with their specific DNA-binding proteins, zinc finger and transcription activator-like repeats. Clustered regularly interspaced short palindromic repeats (CRISPR-Cas9) is directed to the specific genomic site with the help of the protospacer adjacent motif (PAM) in red and the sequence of the single guide RNA (sgRNA) in blue. Once the specific genomic site is recognized, the catalytical subunits, Flavobacterium okeanokoites type IIS restriction enzyme (FokI), or CRISPR-associated protein 9 (Cas9) induces a double strand brake (DSB) and can be repaired either by non-homologous end joining that can lead to insertion/deletion or homology-directed repair with the help of DNA template that allows the introduction of specific mutations. (B) Catalytically inactive Cas9 or dead Cas9 (dCas9) fused to a deaminase (e.g., cytidine or adenine) can result in the conversion of cytosine–guanine base pairs to thymine–adenine or vice versa, without the need of a DSB. A = adenine, T = thymine, C = cytosine, G = guanine, U = uracil, I = inosine. (C) An additional variation of dCas9 fused to a transcriptional activator/repressor can induce transcriptional activation/repression of the targeted gene.