Figure 1. Extended tag:anti-tag pairing prevents RNA cleavage by Cas13a.
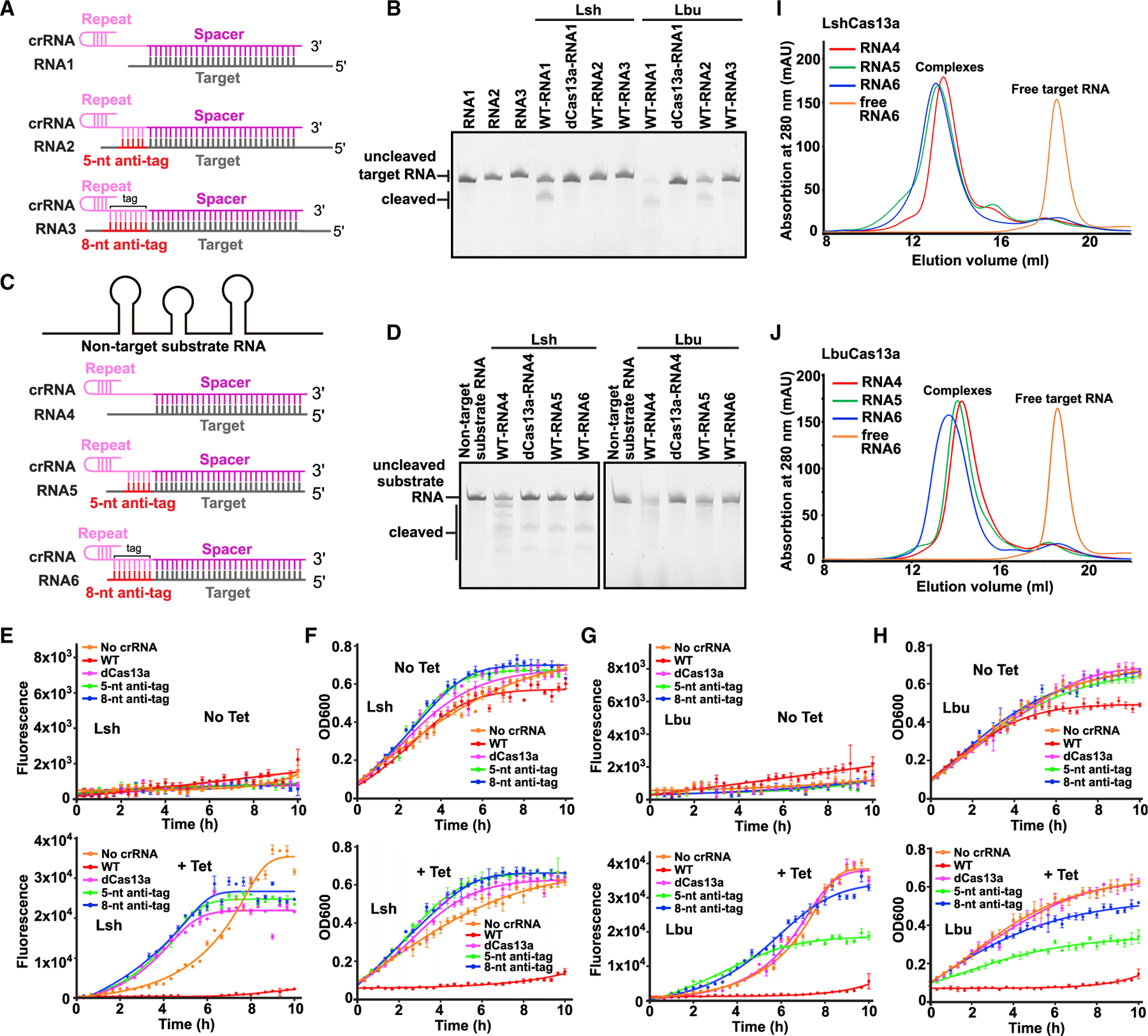
(A and C) Schematic of target RNAs designed for in vitro cis-RNase and trans-RNase cleavage assays. The spacer and repeat of crRNA are shown in magenta and pink, respectively. The target segment is shown in gray, with anti-tag indicated in red.
(B and D) In vitro cleavage assays by LshCas13a and LbuCas13a monitoring substrate RNA degradation on formation of target RNA (B) and non-target substrate RNA (D) ternary complexes in the presence or absence of anti-tag. Two reported dead mutations, LshCas13a-R1278A and LbuCas13a-R472A/H477A, are used as negative controls and labeled as dCas13a. The sequence and schematic of crRNA and target RNAs are shown in (A), (C), and Figures S1B and S1C.
(E and G) Quantification of EYFP mRNA knockdown by LshCas13a (E) and LbuCas13a (G) in E. coli cells. EYFP mRNA contains a 28 nt segment complementary to spacer followed by an 8 nt segment with or without anti-tag. Transcription of EYFP mRNA was induced by tetracycline. Error bars represent standard error of mean (SEM) of three biological replicates.
(F and H) Effect on growth rate of E. coli cells upon EYFP mRNA interference by LshCas13a (F) and LbuCas13a (H). Error bars represent SEM of three biological replicates.
(I and J) Elution profiles run from a Superdex 200 10/300 GL size exclusion column in the presence or absence of anti-tag, showing that anti-tag has no effect on target RNA loading and formation of LshCas13a-crRNA-target RNA (I) and LbuCas13a-crRNA-target RNA (J) ternary complexes.
See also Figure S1.
