Figure 4. Comparison of HEPN pocket alignments and global folds between ternary complexes involving bound anti-tag (LshCas13) and target (LbuCas13) RNAs.
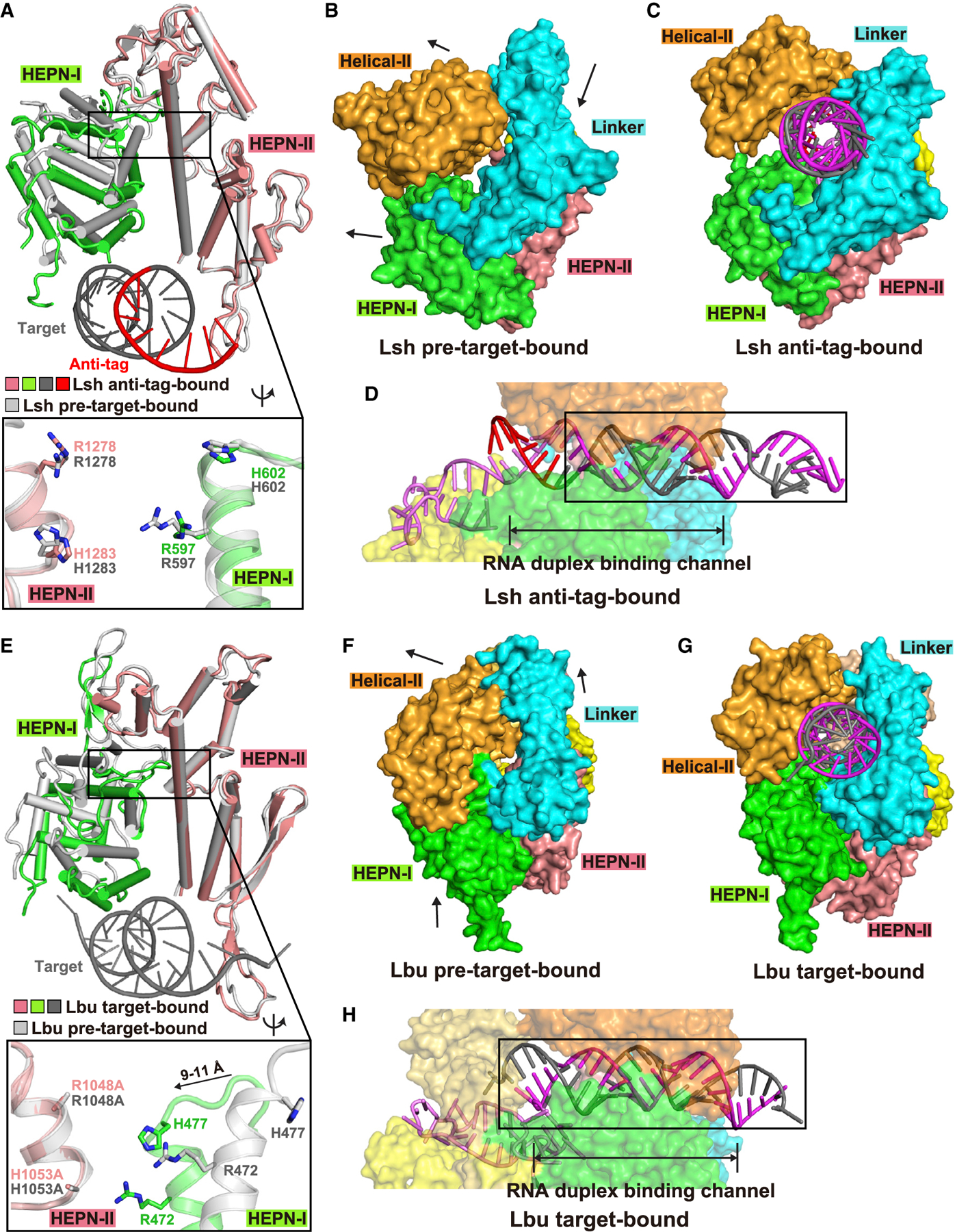
(A) Structural comparisons of two HEPN domains of LshCas13a in anti-tag-bound (HEPN-I in green, HEPN-II in salmon, target segment in gray, and anti-tag in red) and pre-target-bound (in silver) states by superposing the HEPN-II domains. For simplicity, only target RNA is shown. Comparison of the positioning of the four catalytic residues from the pair of HEPN domains is shown in the zoomed-in segment (inset).
(B and C) Surface of LshCas13a showing domain rearrangements to generate crRNA:target RNA duplex binding channel from pre-target-bound (B) to anti-tag-bound (C) states. Black arrows in (B) show the directions of domain movements on ternary complex formation with anti-tag RNA.
(D) Surface views of the interfaces between crRNA-bound LshCas13a and duplex formed by bound anti-tag RNA. The guide:target duplex segment is boxed.
(E) Structural comparisons of two HEPN domains of LbuCas13a in target-bound (HEPN-I in green, HEPN-II in salmon, target segment in gray) and pre-target-bound (in silver) states by superposing the HEPN-II domains (Liu et al., 2017a). For simplicity, only target RNA is shown. Double mutation R1048A/H1053A was used for structural studies of target RNA-bound LbuCas13a. Comparison of the positioning of the four catalytic residues from the pair of HEPN domains is shown in the zoomed-in segment (inset).
(F and G) Surface of LbuCas13a showing domain rearrangements to generate crRNA:target RNA duplex binding channel from pre-target-bound (F) to target-bound (G) states. Black arrows in (F) show the directions of domain movements on ternary complex formation with target RNA.
(H) Surface views of the interfaces between crRNA-bound LbuCas13a and duplex formed by bound target RNA. The guide-target duplex segment is boxed.
See also Figure S4.
