Figure 5. Conformational changes in LshCas13 upon anti-tag RNA loading.
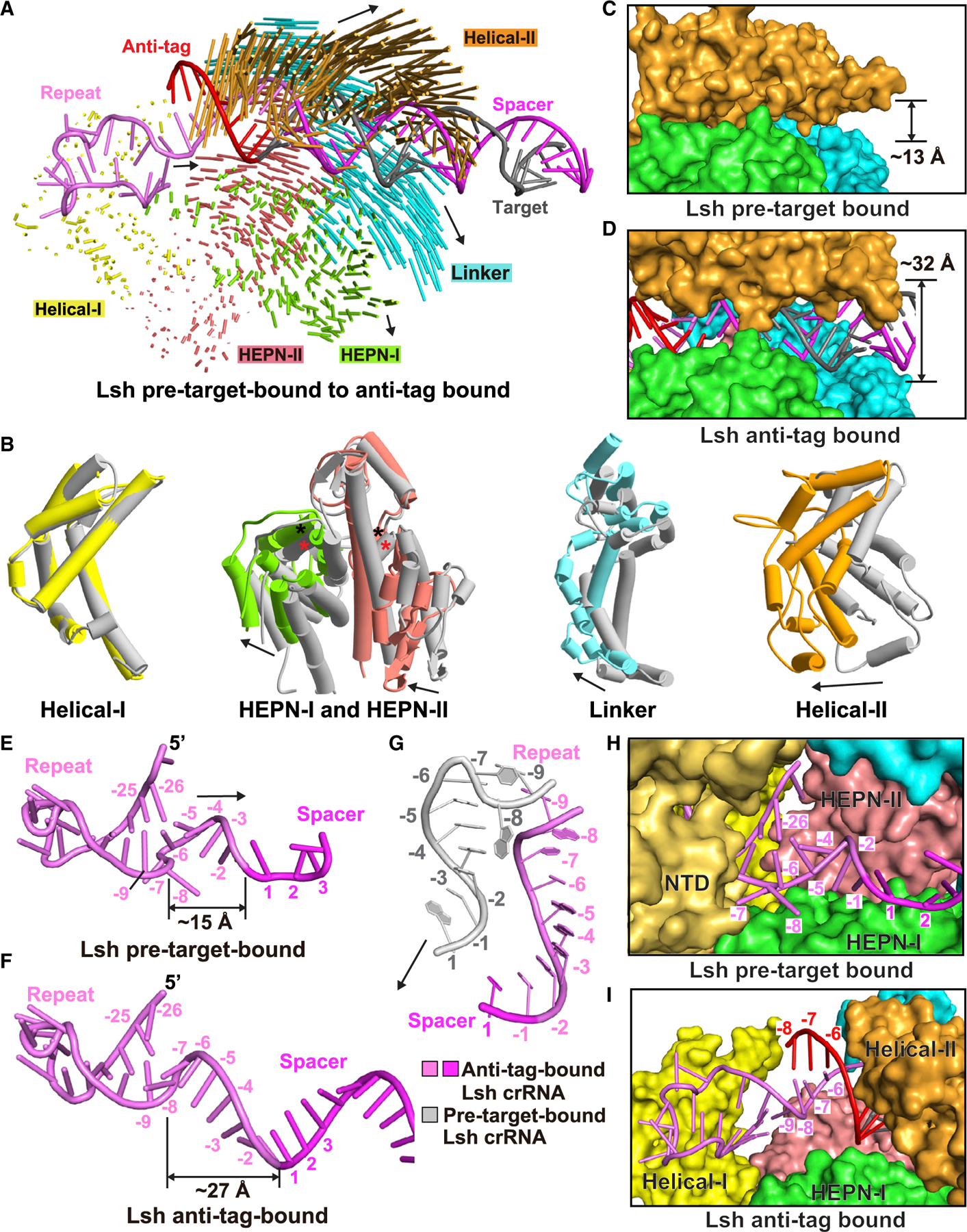
(A) Structural comparison between LshCas13a-crRNA binary complex and LshCas13a-crRNA-anti-tag RNA ternary complex. Vector lengths correlate with the domain motion scales. Arrows show the directions of domain movement from pre-target-bound to anti-tag-bound states.
(B) Structural comparison of the Helical-I, HEPN-I, HEPN-II, Linker, and Helical-II domains between LshCas13a-crRNA binary (in silver) and LshCas13a-crRNA-anti-tag RNA ternary (in color) complexes. Arrows indicate the domain movements. The key catalytic residues from the pair of HEPN domains are indicated by black (anti-tag-bound) and red (pre-target-bound) asterisks, respectively.
(C and D) Binding with target RNA harboring anti-tag widens the guide:target duplex-binding channel on proceeding from LshCas13a-crRNA binary complex (C) to the LshCas13a-crRNA-anti-tag RNA ternary complex (D).
(E–G) Architectures of crRNA in LshCas13a-crRNA binary (E) and LshCas13a-crRNA-anti-tag RNA ternary (F) complexes. The details of tag region are shown in (G), with anti-tag-bound crRNA in color and pre-target-bound crRNA in silver.
(H and I) Comparisons of the tag region of crRNA in LshCas13a-crRNA binary (H) and LshCas13a-crRNA-anti-tag RNA ternary (I) complexes. The Helical-II domain, which interacts with and covers the tag region in LshCas13a-crRNA complex, is hidden in (H) to show the position of tag region.
See also Figure S4.
