Figure 6. Structural comparison between target-bound (LbuCas13a) and anti-tag-bound (LshCas13a) ternary complexes.
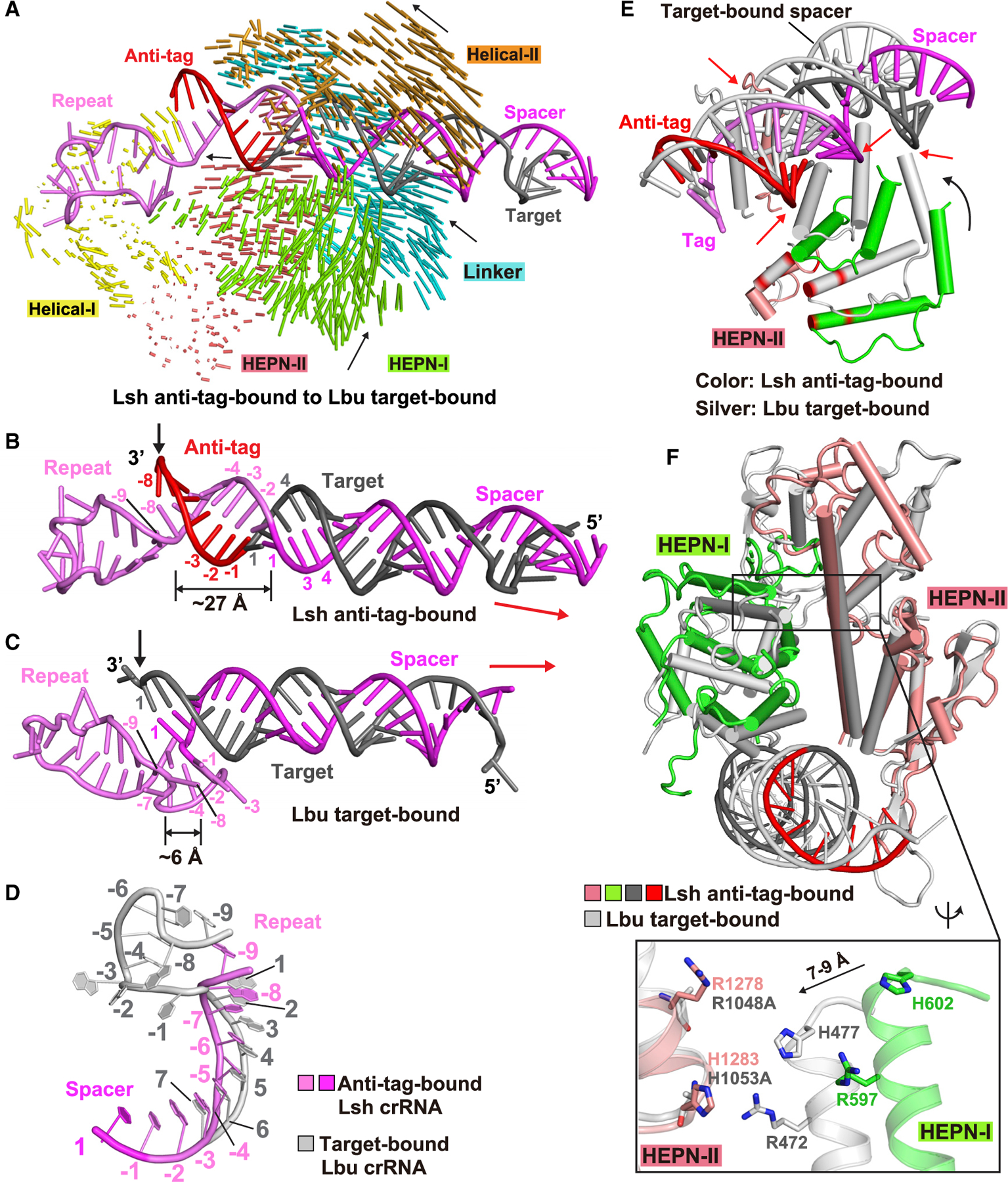
(A) Structural comparison between LshCas13a-crRNA-anti-tag RNA (this study) and LbuCas13a-crRNA-target RNA (Liu et al., 2017a) ternary complexes. Vector lengths correlate with the domain motion scales. Arrows show the directions of domain movement from anti-tag-bound to target-bound states.
(B–D) Architectures of crRNA in LshCas13a-crRNA-anti-tag RNA (B) and LbuCas13a-crRNA-target RNA (C) ternary complexes. The positions of 3′ end of target RNAs are indicated by black arrows. The extension directions of RNA duplexes are indicated by red arrows. The details conformational changes of the tag region are shown in (D), with anti-tag bound crRNA in color and target-bound crRNA in silver.
(E) Superposition of LshCas13a-crRNA-anti-tag RNA (in color) and LbuCas13a-crRNA-target RNA (in silver) ternary complexes with the focus on HEPN-I and HEPN-II domains. The key catalytic residues in HEPN domains are highlighted in red. The black arrow indicates the movements of HEPN-I domain toward the HEPN-II domain from anti-tag-bound to target-bound states. The red arrows indicates the steric clashes between crRNA:anti-tag RNA duplex and the HEPN domains in target-bound state, indicating that the formation of tag:anti-tag RNA duplex prevents the movements of HEPN domains to generate a competent composite catalytic pocket.
(F) Structural comparisons of two HEPN domains LshCas13a in anti-tag-bound (HEPN-I in green, HEPN-II in salmon, target segment in gray, and anti-tag in red) and LbuCas13a in target-bound bound (in silver) states by superposing the HEPN-II domains. For simplicity, only target RNA is shown. Comparison of the positioning of the four catalytic residues from the pair of HEPN domains is shown in the zoomed-in segment (inset).
See also Figure S4.
