Figure 7. Inhibition of RNA cleavage by blockage of conformational changes in tag segment.
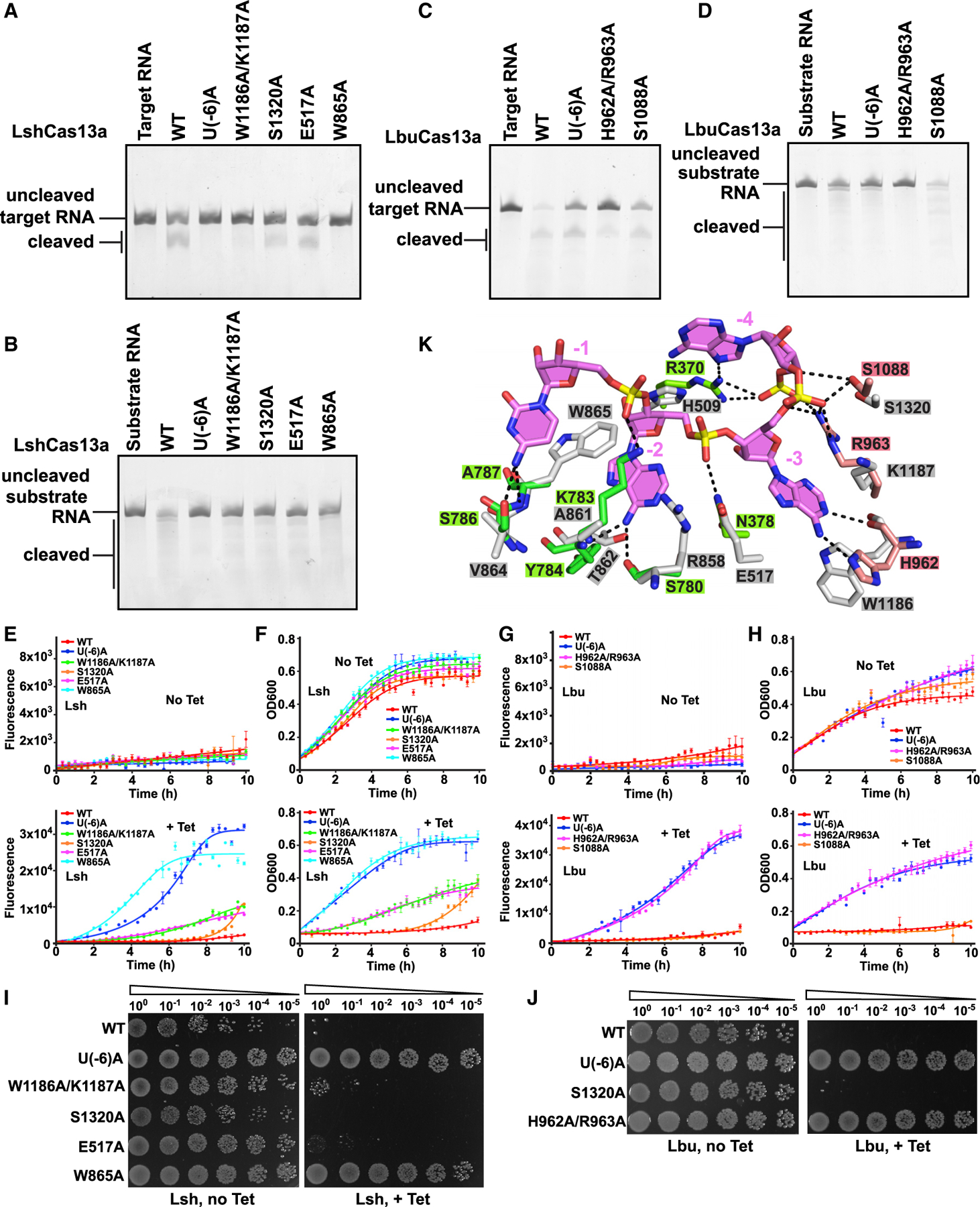
(A and B) Cleavage assays monitoring degradation on target RNA (A) and substrate RNA (B) with Lsh-crRNA or LshCas13a mutants.
(C and D) Cleavage assays by LbuCas13a monitoring degradation on target RNA (C) and substrate RNA (D).
(E and G) In vivo RNA knockdown assays of indicated E. coli strains expressing various LshCas13a (E) and LbuCas13a (G). Error bars represent SEM of three biological replicates.
(F and H) Liquid growth assays for LshCas13a (F) and LbuCas13a (H) illustrating growth defects upon Cas13a-dependent RNA interference. Error bars represent SEM of three biological replicates.
(I and J) Plasmids interference assays for E. coli strains co-transformed with various LshCas13a (I) or LbuCas13a (J) plasmids as well as a tet-induced EYFP plasmid. Ten-fold serial dilution of cells as indicated were spotted onto plates.
(K) Recognition of crRNA 3′-flank (positions −3 to −1) by LbuCas13a (HEPN-I in green, HEPN-II in salmon, and crRNA tag segment in violet) and modeled LshCas13a (in silver).
See also Figure S4.
