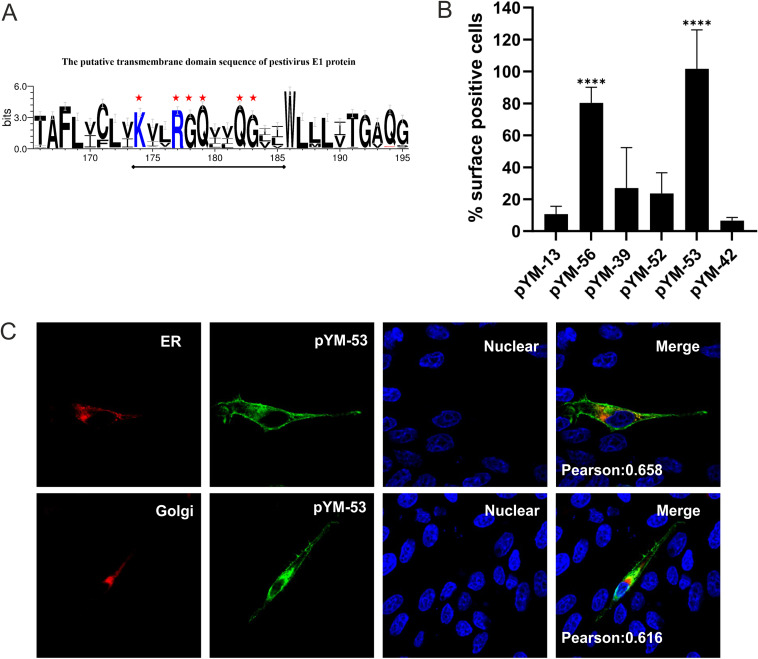
FIG 4.
Six polar residues of E1 TM domain are important for ER retention of E1. (A) Conservation of amino acid sequences in the putative TM region of pestiviral E1. The sequence logo was generated by WebLogo 3 web application (http://weblogo.threeplusone.com/create.cgi) and demonstrates the alignment of 68 pestivirus E1 sequences throughout pestiviral species A to K in one-letter code. The size of the letters in the sequence logo corresponds to the degree of conservation. Six highly conserved polar residues in the middle of the sequence are highlighted with red asterisks. For not fully conserved residues, the height of symbols within the stack represents the relative frequency of each amino acid at the corresponding position. (B) Cell surface expression of HA-tagged E1 mutants analyzed by flow cytometry. The RK-13 cells were transfected with the given expression plasmids and analyzed as described in legend to Fig. 3. The plasmids used include pYM-13 (HA-E1), pYM-56 (HA-E1ecto-VSV-GTMD), pYM-39 (HA-E1 with K174A and R177A), pYM-52 (HA-E1 with K174A, R177A, G182A, and Q183A), pYM-53 (HA-E1 with K174A, R177A, Q178A, G179A, G182A, and Q183A), and pYM-42 (E1 Q182A and G183A). (C) Subcellular localization of the pYM-53-derived HA-E1 mutant containing six mutations (K174A, R177A, G178A, Q179A, Q182A, and G183A) in the TM domain analyzed via coexpression with pDsRed-ER or pDsRed-Golgi, respectively. At 24 h posttransfection, the cells were fixed by 4% PFA, permeabilized with 0.05% Triton X-100, and stained with specific antibodies against HA (green). Compartments (ER or Golgi) are indicated in red. Nuclei were stained with DAPI (blue).