Fig. 2. Probability of symptomatic and severe DENV2 infection by prior DENV and ZIKV infection histories and preexisting antibody titers, 2019–20.
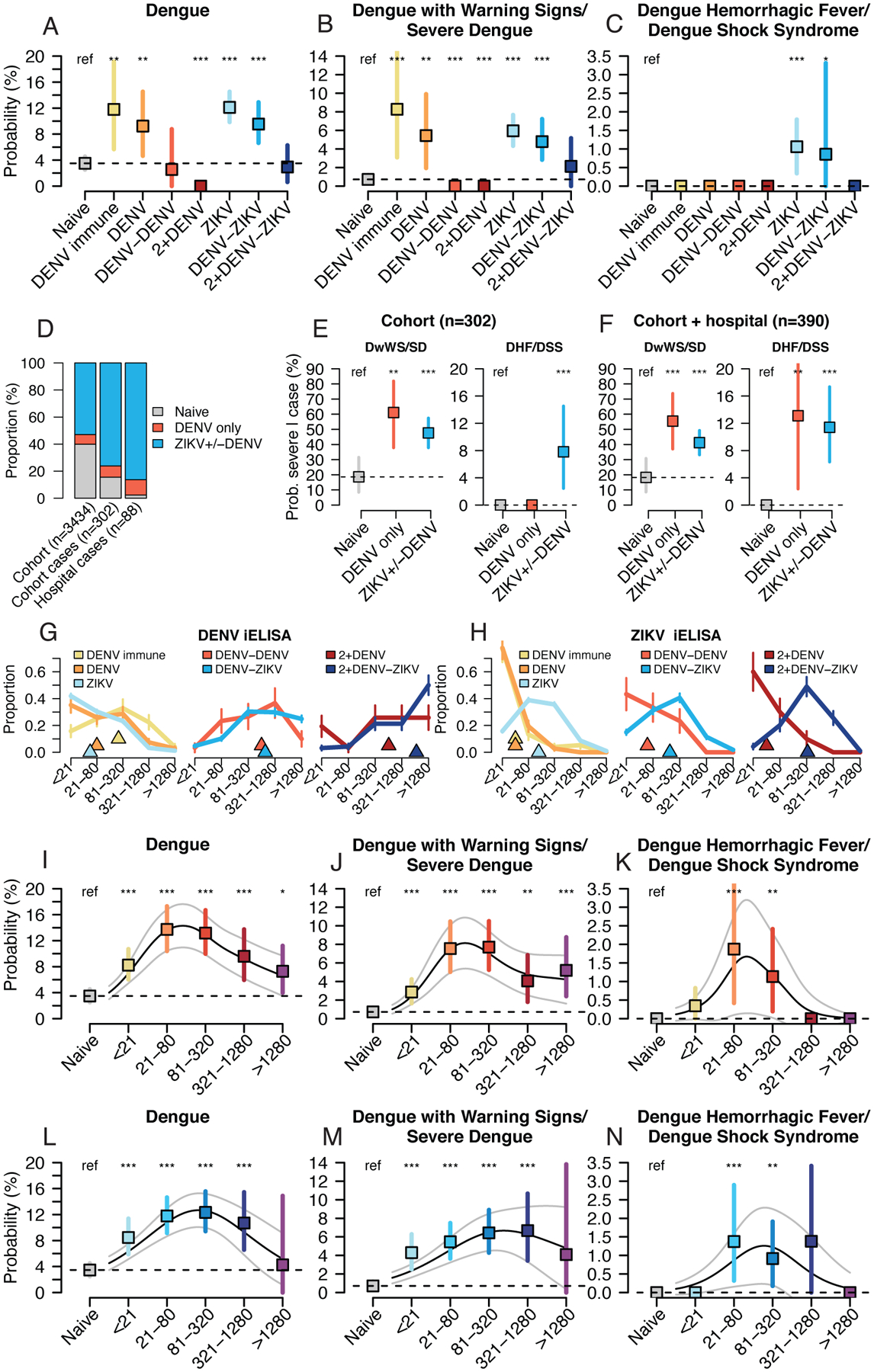
Log-binomial generalized linear models (GLMs) were used to estimate the probability dengue disease in 2019–20 in the cohort study by DENV and ZIKV infection history (A,B,C), preexisting DENV iELISA titers (I,J,K), or preexisting ZIKV iELISA titers (L,M,N), with bootstrap resampling (n=10,000) to construct 95% confidence intervals. Continuous relationships between titers and disease were modeled with log-binomial generalized additive models (GAMs, black lines show probabilities, grey lines show 95% confidence intervals). (D) Infection histories for children in the cohort, cohort dengue cases, and hospital study dengue cases. Differences in the proportion with histories of prior ZIKV+/−DENV or DENV only were tested with a Kruskal-Wallis rank sum test. Probability of severe dengue disease among confirmed dengue cases in cohort (E) or cohort and hospital studies (F) by infection history, estimated using logistic regression. Bootstrap resampling (n=10,000) was used to construct 95% confidence intervals. DENV iELISA (G) and ZIKV iELISA (H) titer distributions for cohort participants in 2019–20 by DENV and ZIKV infection history (triangles show median values, vertical bars show +/−1 standard deviation). All models were adjusted for age and sex, and probabilities are shown for an average study participant (male, age 8). P-values (*, <0.05; **, <0.01; ***, <0.001) indicate significantly different probability estimates from the Naïve group.
