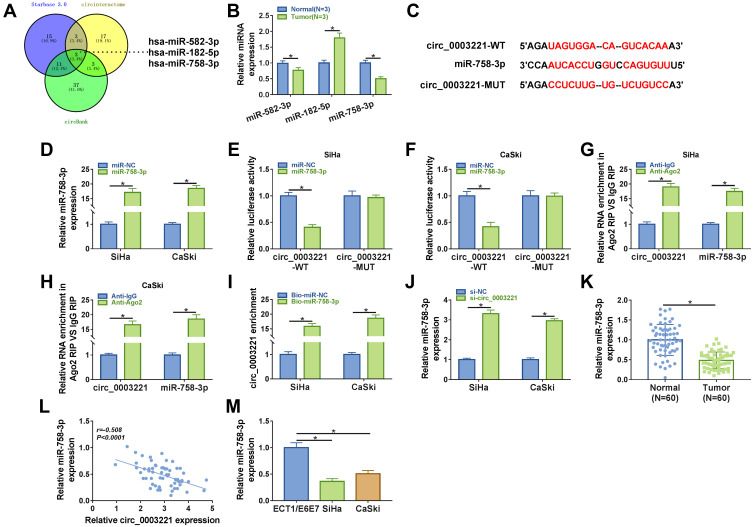
Figure 3.
MiR-758-3p was a direct target of circ_0003221. (A) 3 miRNAs (has-miR-582-3p, has-miR-182-5p, and has-miR-758-3p) that might be sponged by circ_0003221 were predicted by three databases (Starbase 3.0, Circinteractome and circBank). (B) The expression of miR-582-3p, miR-182-5p, and miR-758-3p was measured in normal tissues and cervical cancer tissues by qRT-PCR. (C) The binding sequences between miR-758-3p and circ_0003221 were presented. (D) The expression of miR-758-3p was detected by qRT-PCR in SiHa and CaSki cells transfected with miR-NC or miR-758-3p. (E–I) Dual-luciferase reporter assay, RIP assay and RNA pull-down assay were conducted to confirm the interaction between miR-758-3p and circ_0003221. (J) The level of miR-758-3p was detected by qRT-PCR in SiHa and CaSki cells transfected with si-NC or si-circ_0003221. (K) The expression of miR-758-3p was detected by qRT-PCR in normal tissues and cervical cancer tissues. (L) The correlation between miR-758-3p and circ_0003221 was analyzed in cervical cancer tissues. (M) QRT-PCR was used to examine the expression of miR-758-3p in ECT1/E6E7, SiHa and CaSki cells. *P<0.05.