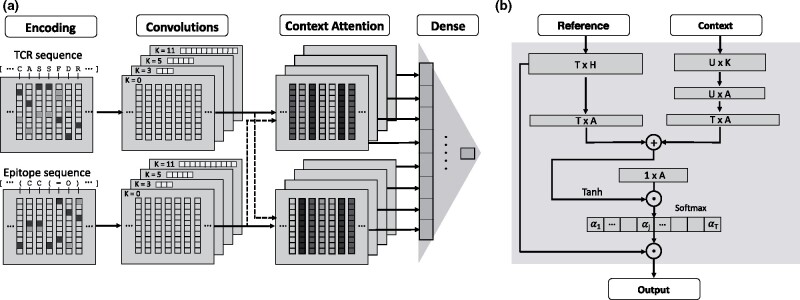
Fig. 1.
Overview of TITAN architecture. (a) Our model ingests a TCR and an epitope sequence, which get encoded using BLOSUM62 for amino acid sequences or learned embeddings for SMILES. Then, 1D convolutions of varying kernel sizes are performed on both input streams before context attention layers generate attention weights for each amino acid of the TCR sequence given an epitope and vice versa. Finally, a stack of dense layers outputs the binding probability. Conceptually, this architecture is identical to the one proposed in Born et al. (2021) (cf. Supplementary Fig. S3) but our visualization here is more fine-grained. (b) The linchpin of the model is the bimodal context attention mechanism. It ingests the convolved TCR and epitope encodings, treating one as reference and the binding partner as context. A series of transformations combines the modalities and yields an attention vector over the reference sequence (driven by the context) that can be overlayed with the molecule like a heatmap