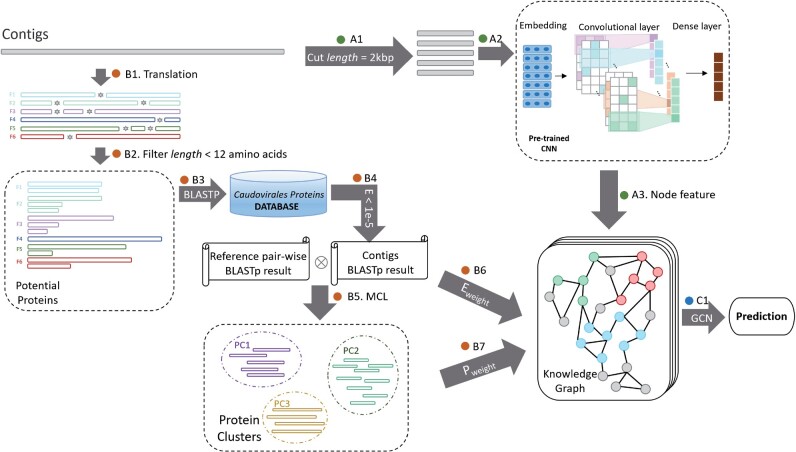
Fig. 1.
The pipeline of PhaGCN. A1: cut the contigs into 2 kbp segments. A2: feature learning from the inputs using CNN. A3: construct nodes using encoded vectors. B1: contig translation using 6 reading frames. B2: filter short translations (12 amino acids). B3: align contigs against reference database using the DIAMOND BLASTP command. B4: choose the best translated frame for the BLASTP result. B5: use the BLASTP result to construct protein clusters. B6 and B7: define edges based on the sum of the Eweight and Pweight. C1: construct the knowledge graph for GCN