Fig. 2.
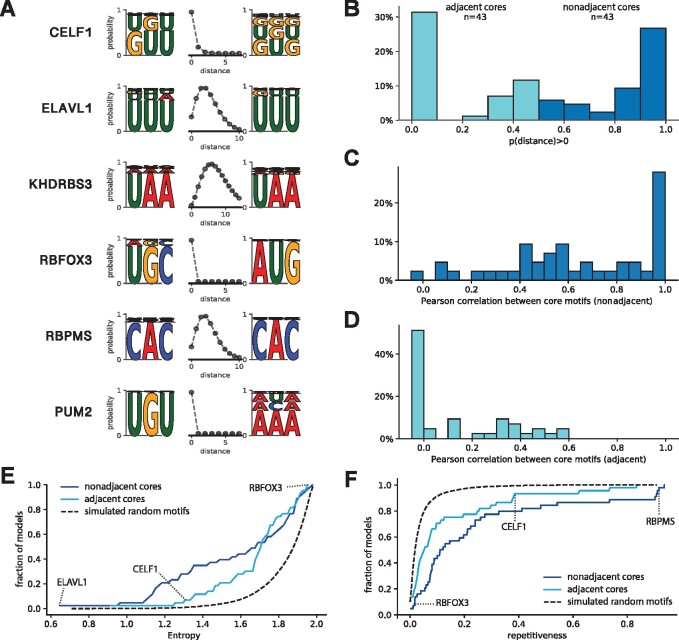
Many RBPs are multivalent, bind low-complexity sequences and often bind two similar motif cores. (A) Examples of motifs that represent a wide range of binding modes, learned by BMF on HTR-SELEX data. When the RBP has a larger motif than allowed by the core size (3 here), the distance between cores is learned to be zero to accommodate a longer binding sequence (e.g. CELF1, RBFOX3 and PUM2). (B) Distribution of the probability of the spacer length between the two motif cores to be above 0. As seen in the examples in A, most RBPs either clearly bind adjacent cores (distance, turquoise) or have a multivalent binding mode with two non-adjacent cores (dark blue). (C) and (D) Similarities between binding preferences of the two cores for RBPs with adjacent cores (turquoise) or multivalent non-adjacent cores (dark blue), according to panel B. (E) Cumulative distribution of the entropy of BMF models for all RBPs in the HTR-SELEX dataset. In general the optimized bipartite motif models have much lower complexity than randomly generated bipartite models (dashed black line). (F) Cumulative distribution of ‘sequence repetitiveness’ of BMF models for all RBPs in the HTR-SELEX dataset. Overall, BMF models are more often repetitive that those of randomly generated bipartite models (dashed black line)