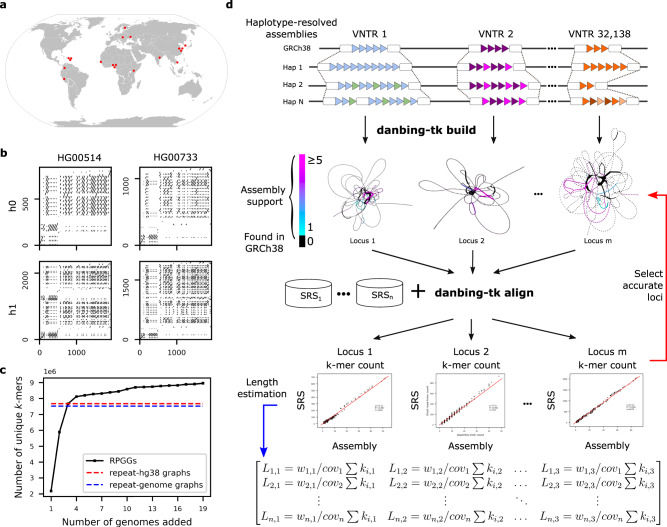
Fig. 1. Sequence diversity of VNTRs in human populations.
a Global diversity of long-read assemblies. b Dot-plot analysis of the VNTR locus chr1:2280569–2282538 (SKI intron 1 VNTR) in genomes that demonstrate varying motif usage and length. c Diversity of RPGG as genomes are incorporated, measured by the number of k-mers in the 32,138 VNTR graphs. Total graph size built from GRCh38 and an average genome are also shown. d Danbing-tk workflow analysis. (top) VNTR loci defined from the reference are used to map haplotype loci. Each locus is converted to a de Bruijn graph, from which the collection of graphs is the RPGG. The de Bruijn graphs shown illustrate sequences missing from the RPGG built only on GRCh38. The alignments may be either used to select which loci may be accurately mapped in the RPGG using SRS that match the assemblies (red), or may be used to estimate lengths on sample datasets (blue). Source data are provided as a Source Data file.