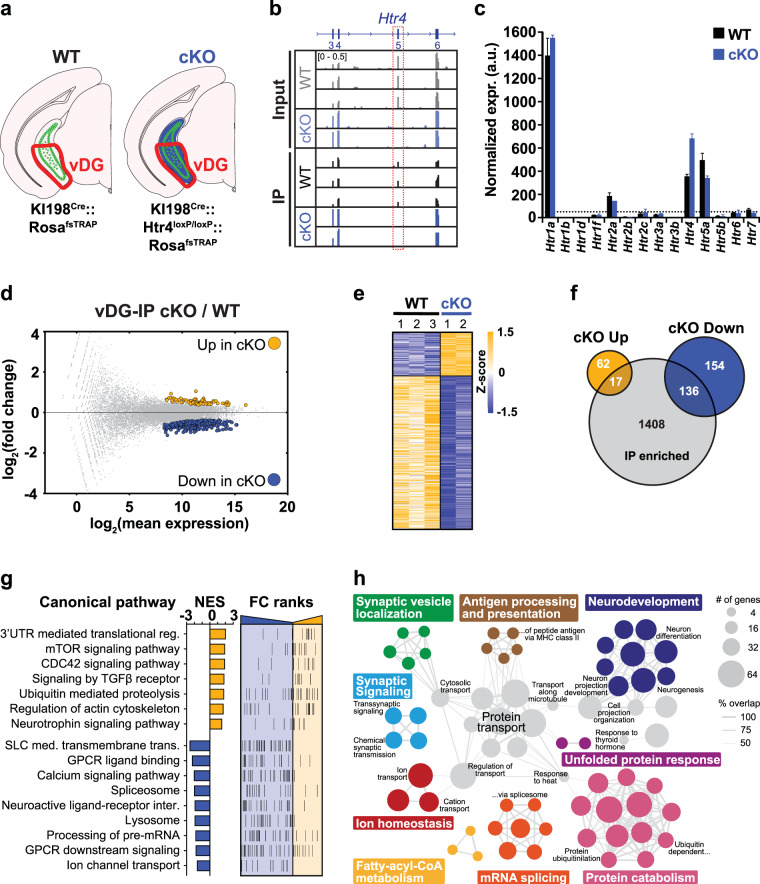
Fig. 4. TRAP profiling of vDG neurons in the absence of 5-HT4R.
a Schematic depicting the cell types targeted for TRAP. Green represents EGFPL10a expressing cells and blue indicates conditional 5-HT4R deletion. The area dissected is outlined in red. b Browser view of mapped reads showing the lack of expression of Htr4 exon 5 in IP and input samples in the cKO. c Bar graph of normalized expression values (mean ± SEM) of all 5-HT receptors (Htrs) in IP samples. Dashed line is drawn at normalized expr. = 50. Only Htr4 has FDR < 0.1. d MA-plot highlighting differentially expressed (DE) genes (FDR < 0.1) between the cKO and WT TRAP mRNA. Complete list of IP DE genes can be found in Table S1. e Heatmap visualization of DE genes between genotypes. f Venn diagram showing the overlap of up- and down-regulated DE genes from e with genes enriched in the KI198Cre -expressing cells (IP enriched, Table S2). g Summary of gene set enrichment analysis (GSEA) performed on the fold change (FC)-ranked gene list (p < 0.05) between cKO and WT IP data sets. The normalized enrichment score (NES) shows the direction of enrichment in the cKO and FC-ranks are arranged from down-regulated (blue) at left to up-regulated (yellow) at right. Complete list of pathways can be found in Table S3. h Summary of gene ontology (GO) analysis of DE from e. All categories shown have FDR < 0.01, full analysis is provided in Table S4.