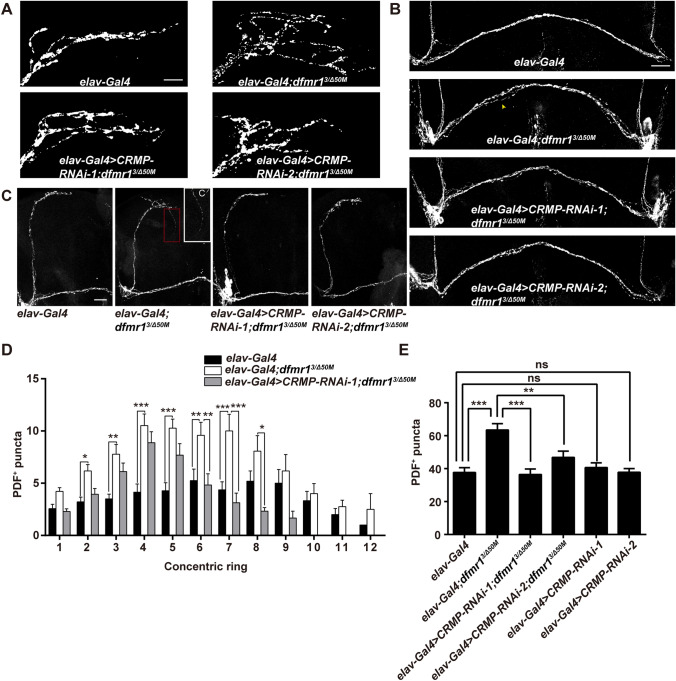
Fig. 2.
Genetic reduction of CRMP rescues structural deficits of LNv neurons in dfmr1 mutants. A Terminals of sLNvs at the dorsal protocerebrum of the depicted genotypes. dfmr1 mutants show increased branching complexity, while knockdown of CRMP with both RNAi transgenes in the nervous system reduces the over-elaborated axonal branches (scale bar, 10 μm). B Representative split POT phenotype (indicated by arrowhead) in the indicated flies. dfmr1 mutants show a POT defasiculation phenotype compared with the wild-type, and reduced CRMP expression rescues the POT splitting (scale bar, 75 μm). C Half brains of adult flies showing collateral branches. C’ Magnification of the collateral branch in a dfmr1 mutant Drosophila. Some of dfmr1 mutants have ectopic collateral branches (indicated by the red dash box), while mutant flies with two CRMP RNAi transgenes display no collateral branching, like wild-type flies (scale bar, 75 μm). D Sholl analysis of the distribution of PDF-reactive puncta throughout the sLNv axonal arbors. dfmr1 mutants harbor increased PDF puncta distributed in each ring, which is rescued by knockdown of CRMP. E Total numbers of PDF-positive (PDF+) puncta throughout the sLNv axonal arbors. Knockdown of CRMP with both RNAi transgenes rescues the significantly elevated total number of PDF puncta in dfmr1 mutants. *P <0.05, **P <0.01, ***P <0.001, two-way ANOVA for C and one-way ANOVA for D. n >20 hemispheres per genotype. Data are represented as the mean ± SEM. ns, not significant.