Abstract
The EPA developed the Adverse Outcome Pathway Database (AOP-DB) to better characterize adverse outcomes of toxicological interest that are relevant to human health and the environment. Here we present the most recent version of the EPA Adverse Outcome Pathway Database (AOP-DB), version 2. AOP-DB v.2 introduces several substantial updates, which include automated data pulls from the AOP-Wiki 2.0, the integration of tissue-gene network data, and human AOP-gene data by population, semantic mapping and SPARQL endpoint creation, in addition to the presentation of the first publicly available AOP-DB web user interface. Potential users of the data may investigate specific molecular targets of an AOP, the relation of those gene/protein targets to other AOPs, cross-species, pathway, or disease-AOP relationships, or frequencies of AOP-related functional variants in particular populations, for example. Version updates described herein help inform new testable hypotheses about the etiology and mechanisms underlying adverse outcomes of environmental and toxicological concern.
Subject terms: Data integration, Genetic databases, Protein databases
| Measurement(s) | adverse outcome pathway • gene interactions • Orthologous Gene • chemical gene interactions • molecular pathway • disease gene associations • SNP |
| Technology Type(s) | digital curation |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.14737557
Background & Summary
There is a need for approaches to understand the biological mechanism of adverse outcomes and human variability in response to environmental chemical exposure. A recent legislation, the Frank R. Lautenberg Chemical Safety for the twenty-first Century Act of 20161, requires the US Environmental Protection Agency to evaluate new and existing toxic chemicals with explicit consideration of susceptible populations of all types (life stage, exposure, genetic, etc.). In addition, on September 10, 2019, EPA Administrator Andrew Wheeler signed a directive that prioritizes efforts to reduce animal testing. In response to this directive, the EPA has developed a 2019 Strategic Plan to Promote the Development and Implementation of Alternative Test Methods Strategies (or New Approach Methodologies (NAMs)) per TSCA Section 4(h)(2)(C). The EPA Adverse Outcome Pathway Database (AOP-DB) is a decision support tool developed by the EPA’s Center for Public Health and Environmental Assessment, which contributes to NAMs (e.g. computational toxicology tools) used for TSCA. The EPA Adverse Outcome Pathway Database (AOP-DB) is a database resource that combines different data types (AOP, gene, chemical, disease, pathway, orthology, and ontology) to characterize the impacts of chemicals to human health and the environment2, and for the characterization of human genetic susceptibility for the purpose of human health risk assessment3. The AOP-DB was originally developed with the primary aim of integrating AOP molecular target information with other publicly available datasets and related toxicological data, to facilitate computational analyses of AOP information. Near term goals for use of the AOP-DB are to address the biological and mechanistic aspects of alternative test methods in terms of the adverse outcome pathway construct to facilitate Integrated Approaches to Testing and Assessment (IATA) for regulatory purposes4–6.
Here we present an updated version of the database, AOP-DB v.2, which includes an increased number of adverse outcomes and corresponding key events derived from updated feeds from the AOP-Wiki 2.0 (https://AOP-Wiki.org/), as well as updated chemical, disease, tissue, individual and population level data and ontology information. In this second iteration of the database, we update all code and previously described data from AOP-DB version 1, as well as provide integration with three new data areas: tissue, individual and population level data. We discuss the collaborative semantic mapping efforts for AOP-DB data, and highlight the AOP-DB web user interface, which will be deployed to the public in 2021.
Pittman et al.2 presented the first version of the AOP-DB which focused on chemical and species-specific analyses. Mortensen et al.3 developed a computational approach that implements the AOP-DB to integrate mechanistic data associated with an AOP with data capturing human genetic variability and function, for the purpose of characterizing human molecular variation that may impact individual and population level responses to environmental chemicals. The human individual and population level data included in AOP-DB v.2 provide the relevant data sources and organizational structure to envision the approach presented by Mortensen et al.3. We believe that this work represents a first step in organizing a coherent research program in molecular environmental adverse outcomes. Current and future work will focus on the application and interrogation of these data for case study examples, the development of computational, quantitative AOP (qAOP) models for estimation of chemical-MIE protypes, and machine learning methods to understand human susceptibility and variation in functional response that will inform chemical safety assessment.
Methods
Data included in the AOP-DB v.2 represent an aggregation of publicly available sources associated with adverse outcome pathways. All data included in AOP-DB v.2 were acquired or generated as detailed below. Online-Only Table 1 lists the current data sources integrated in the AOP-DB v.2. Table 1 lists the actual record count for integrated data within the AOP-DB v.2. For example, 261 supported AOPs in the AOP-DB v.2, as indicated in Table 1, indicates there are 261 unique, expert-derived AOPs that map to one or more unique gene or protein accession numbers. Similarly, for those 261 AOPs, 1029 chemical stressors map chemical ID to AOP ID, underlining a unique AOP-chemical association. The AOP-DB v.2 contains a full list of AOP stressors obtained and updated from the AOP-Wiki 2.0, which is available and updated as part of the OECD supported AOP Knowledge Base (AOP-KB) through e.AOP.Portal (https://aopkb.oecd.org/).
Online Only Table 1.
Additional data sources included in the AOP-DB.
| Biological Category | Data Source | Description | Processing | URL |
|---|---|---|---|---|
| Gene | NCBI Gene | This source supplies all NCBI entrez genes in the gene info table with associated gene information such as name, symbol, location, etc. | The latest gene_info and gene_history files are pulled from the NCBI GENE FTP. gene_history contains all deprecated entrez which are used to remove entries from the gene_info table. No other processing takes place | ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/ |
| STRING | This source gives protein-protein interaction data for the gene-interactions table. Each record from these networks is stored with an entrez1, entrez2, and an interaction score | STRING files are downloaded (no api.or ftp) and proteins are mapped to entrez via unitprot. (UPKB2entrez). No other processing takes place | http://string-db.org/cgi/download.pl | |
| Taxonomy & Orthology | NCBI Taxonomy | All taxa available from NCBI, including nomenclature info and divisions. This data is used to fill the species_info | The taxonomy data are pulled from the NCBI FTP. No other processing takes place. | ftp://ftp.ncbi.nlm.nih.gov/pub/ |
| Homologene | Constructs and stores putative homology groups and contributes and ortho group number, tax id, entrez id to the homology gene table | The homogy_gene file is pulled from the NCBI FTP. No processing takes place. | ftp://ftp.ncbi.nlm.nih.gov/pub/HomoloGene | |
| KEGG Orthology | This database of functional orthologs contributes ortho group ids, tax ids, and entrez ids describing an orthologous group to the homology gene_table | The latest ortholog data is pulled from the KEGG “R” Package KEGGREST. Kegg Gene IDs are mapped to entrez. No Processing takes place. | http://www.kegg.jp/kegg/rest/keggapi.html | |
| metaPhOrs | This database of phylogeny based orthologs contributes ortho group ids, taxonomy ids, and entrez ids to the homology gene table, describing orthologous groups. | Mappings and ortholog data for a manually curated set of species are pulled from the phylomedb FTP. Only protein mappings from SwissProt and TrEMBL are considered. Protid IDs are mapped to entrez. Where protid2 is unmapped or empty, it is mapped to entrez = 0. No other processing takes place | ftp://phylomedb.org/metaphors/ | |
| AOP | AOP-wiki | This is a collaborative set of AOPs regularly updated with new details or new Adverse Outcome Pathways. This source contributes to the central AOP_info tables and the AOP_gene tables, supplying AOP names, key events, descriptions, and information used to map key events to genes. | The latest aop wiki xml dump is downloaded from the AOPWiki downloads page. No other processing takes place | https://aopwiki.org |
| Chemical | CTD | This source is a manually curated database of chemical information, including many modules. The module of interest for the AOPdb is the chemical gene interactions module, which contributes chemical names and ids to chemical info, as well as the chemical gene interactions with contextual information to the chemical gene table. | The latest chemical tables and chemical gene interactions are downloaded from CTD website. No Processing takes place. | http://ctdbase.org/downloads/ |
| AOP-wiki | In addition to being the source of AOPs for the AOPdb, this source also adds chemical stressors related to the MIE of each AOP. This data contributes chemical names, as well is DTXSIDs, casrns, or other chemical ID when available. | Latest AOPWiki XML dump is downloaded. All aop, stressor, and key event information is merged together. No other processing takes place | https://aopwiki.org | |
| ToxCast | This is a collection of high-throughput screening assays for chemicals that contributes assay identification information and assay context information as well as gene target information in the form of entrez ids. | ToxCast Assay data is loaded from a static file. No processing takes place | ftp://newftp.epa.gov/comptox/High_Throughput_Screening_Data | |
| Pathway | KEGG Pathways | This source is a collection of biological molecular interaction pathways that supplies entrez ids and pathway names and ids, linking gene components to the pathways in which they are involved. | Pathway data is accessed through the KEGG API, KEGGREST (version 1.24.0). KEGG gene IDs are mapped to Entrez ID. Entries without an Entrez are dropped. No other processing takes place. | http://www.kegg.jp/kegg/rest/keggapi.html |
| Reactome | This curated and peer-reviewed source of molecular pathways supplies entrez ids and their linked pathways to the pathway_gene table of the AOPdb | Reactome data is loaded, and Tax_ids are mapped using species name and species ID. No other processing takes place. | http://www.reactome.org/pages/download-data | |
| ConcensusPathDB | This source brings together pathway and interaction data from 32 public resources and supplies entrez ids and pathway ids that link genes to biological pathways for the pathway gene table | CPDB data is loaded from a static file. Tax_ids are added by mapping entrez ID to species ID. No other processing takes place. | http://consensuspathdb.org/ | |
| Disease | DisGeNET | This database compiles different data, both curated and inferred from models, and supplies multiple downloadable tables relating genes and variants to the diseases in the database. The AOPdb uses DisGeNET’s gene-disease association table, adding all fields to the disease-gene table. These include disease name and id, entrez id, and a score for the association based on its sources. | The latest disese gene associations are downloaded via the Disgenet website. No other processing takes place. | http://www.disgenet.org/downloads |
| Ontology | NCBI Gene | In addition to being a source of taxonomy info and gene info, NCBI Gene supplies gene ontology information. This supplies gene ontology terms and any related entrez ids to the GO gene table. | The latest gene2go (gene GO mapping file) is downloaded from the NCBI FTP site. No other processing takes place. | ftp://ftp.ncbi.nlm.nih.gov/gene/DATA |
| Tissues | HumanBase | This API is used to pull tissue specific gene interaction network from HumanBase. The data imported into the tissue networks table in the AOPdb include entrez1 and entrez2 fields to construct edges, as well as a probability score indicating the strength of the modeled gene interaction. | Tissue networks are independently retrieved through the HumanBase API for all genes and all tissues. Tissue-gene combinations without edges are dropped. Genes are compared to NCBI’s list of deprecated and entrez ID is updated. If an edge has a deprecated gene it is dropped, if it is changed the tissue_networkgene is updated. No other processing takes place. | https://hb.flatironinstitute.org/api/ |
| Haplotypes | 1000 Genomes | This is a collection of variant data for individuals from a multitude of populations. This source contributes snp frequencies for each function snp in the snps table for each of 5 1000 Genomes major populations. | 1000 Genomes Phase 3 static VCF files are downloaded and stored locally. Using VCF tools we filter the data set to SNPs retrieved from GWAS and GTEX. No other processing takes place. | |
| Ensemble | This API, allowing access to ensembl’s gene and variant information, is used to get genotype data for each individual sample from the 1000 Genomes project. These data are used to construct haplotypes for each AOP and find differences in haplotype frequencies within and between populations. | Ensembl Regulatory regions are loaded from a static file and filtered by AOP-DB SNPs. Genotype, minor allele, variant id are then downloaded for each loci using the Ensembl REST API. No other processing takes place. | https://rest.ensembl.org/ | |
| GWAS Catalog | This is a source used to filter SNPs into snps of interest for variant analysis in different populations. Functional snps are specifically targetd. It, along with GTEx, supplies refsnp ids for these variants as well as contextual information. | SNPs are pulled from the static file GWAS Catalog 1.0.2. No other processing takes place. | https://www.ebi.ac.uk/gwas/ | |
| GTEx | This is a source used to filter SNPs into snps of interest for variant analysis in different populations. Functional snps are specifically targetd. It, along with GWAS Catalog, supplies refsnp ids for these variants as well as contextual information. | SNPs are pulled from the static file Gtex V7 Single Tissue cis eQTL. No other processing takes place. | https://www.gtexportal.org/home/ |
Table 1.
AOP-DB tables with Summary Count information.
| Data Table | Data Type | Count |
|---|---|---|
| Gene Info | Unique gene IDs | 24609215 |
| Gene Interactions | Pairwise gene interaction scores | 4.83E + 08 |
| Species Info | Entrez-supported organisms | 26554 |
| Homology Gene | Orthologous Groups | 64930 |
| Taxa supported by ortho groups | 605 | |
| AOP Info | Supported AOPs | 261 |
| AOP gene | AOP gene associations | 758 |
| Chemical Info | CTD chemicals | 170956 |
| Chemical gene | chemical-gene associations | 1206437 |
| AOP Stressor | DTX-AOP associations | 654 |
| ToxCast Assays | Assays | 406 |
| Pathway Gene | Pathways | 110889 |
| Pathway-entrez links | 6412846 | |
| Disease Gene | Diseases | 24166 |
| Disease-gene associations | 628685 | |
| GO Gene | GO terms | 26739 |
| GO-gene associations | 1698353 | |
| Tissue Networks | tissues | 145 |
| edges | 16957011 | |
| SNPS | refsnp_ids | 2464 |
| AOP gene associations | 5217 | |
| SNP frequencies | populations | 5 |
| Haplotypes | samples | 2504 |
Methods and included data sources presented here reflect the AOP-DB v.2 as of May 5th, 2021. In addition to regular updates to the AOP-DB, we anticipate that additional data will be added to the database over time. The addition of new data will be announced via the AOP-DB web user interface https://aopdb.epa.gov/, on the AOP-DB ‘Home’ page https://www.epa.gov/healthresearch/adverse-outcome-pathway-aop-database, and the CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/), hereafter referred to as the “Dashboard”, on the ‘News’ page, (https://comptox.epa.gov/dashboard/news_info) and ‘Downloads’ (https://comptox.epa.gov/dashboard/downloads) pages, as appropriate. The third-party data sets included in the AOP-DB v.2, and additional details of methods used for any modifications performed in the integration process, are described briefly below and with full details in Fig. 1 and Online-only Table 1.
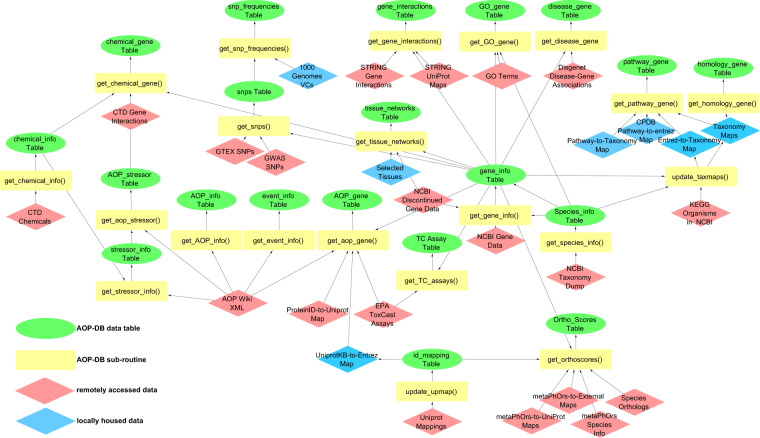
Fig. 1.
AOP-DB Data Structure. Green, Ovals indicate data tables in the AOP-DB SQL relational database; Blue, Diamonds indicate local, post-processing files necessary, where modified data are stored; Yellow, Rectangles indicate corresponding subroutines necessary to process source data; and Red, Diamonds indicate publicly available, third party source data included in AOP-DB v.2. Arrow edges indicate foreign key relationships.
AOP-DB v.2
Data selection and collection methods match closely with those used for AOP-DB v.12. AOP-DB v.2 implements the AOP set available with the AOP-Wiki 2.0 (https://aopwiki.org/). AOP-DB v.12 links AOPs to gene and protein accession numbers manually using the concept of the Event-Component using selected gene and protein ontologies, according to the methods described in Ives et al.7. The AOP-Wiki 2.0 reports the results of an automated ontology mapping process, making automated AOP-gene association updates possible in AOP-DB v.2. AOP-DB v.2 tables are created using MySQL (version 5.7.25) and updated using in-house Perl (version 5.26.3) and Python (3.6) scripts. Data is downloaded and processed from all third-party sources using R (version 3.6.0) and Python (3.6). The AOP-DB v.2 data structure, as described in Fig. 1, illustrates the data integration of the central AOP-gene data with other third-party sources. Figure 1 also indicates how data are integrated across data sets, where subroutines are needed, and local files are stored. The AOP-gene table links chemical-gene (CTD8), biological pathway (ConsensusPathDB9,10, KEGG Pathways11–13, Reactome14), disease-gene (DisGenet v.6.015), species homology (Homologene16), ToxCast Assay target information17–20, tissue-gene mapping (HumanBase21, and population Single Nucleotide Polymorphism (SNP) frequency data for functionally relevant AOP gene targets (Ensembl22,23; GWAS24; GTex25–27; 1KGenomes26,28). The AOP-DB v.2 SQL database schema is included in Supplemental Fig. 1.
AOP-Wiki XML import, and mapping of molecular identifiers
AOP-DB v.2 has updated the primary source of AOP information, and now implements the AOP-Wiki 2.0 XML dump, which is updated quarterly. This dump includes all stressor, event, status, and description data associated with AOP’s from the AOP-Wiki 2.0 in XML format. We used the XML package (version 3.98-1.2) in conjunction with xPath (version 3.1) notation to parse the AOP-Wiki 2.0 XML into tabular data for import into the AOP-DB v.2, in order to create a data frame with essential columns. Once organized into tabular format, this processed XML data is separated into AOP_info, AOP_gene, AOP_stressor, stressor_info, and event_info tables. Individual subroutines automate and validate the accuracy of the parsed data at each stage. The AOP_stressor and stressor_info tables, due to somewhat loosely structured submission structure, require manual processing and mapping of chemical ids to chemicals without mapping information, discussed below.
To create the link between AOP’s from the AOP-Wiki 2.0 and gene identifiers in the AOP-DB v.2, which are not supplied in the AOP-Wiki 2.0 XML directly, we map key event information within each AOP containing a biological object, where a biological object is comprised of three ontological components: Process; Object; and Action. When the object term contains a protein ontology (PR) value, this value is considered a “molecular identifier” and is used to map that key event to a corresponding gene identifier (e.g. Entrez, UniProt, etc.).
AOP-Wiki Stressor tables
After the AOP-Wiki XML is parsed, the AOP-DB stressor tables undergo an additional processing step. The AOP-Wiki XML contains stressor_name, user_term, stressor_id and Chemical Abstract Service Identifier (CASRN). Stressors entered into the AOP-Wiki can include a link to chemical stressors, via the DSSTox Substance Identifier (DTXSID), which maps the stressor to substances registered in the DSSTox database29. The chemical DTXSID, a unique substance identifier, provides a link to the Dashboard using the process described in Williams et al.30. When no DTXSID is provided for stressors imported from the AOP-DB, manual curation to the Dashboard has been performed on individual substances, on a substance-by-substance basis and using available identifiers (e.g. CAS Registry Numbers and chemical names) according to the process described in Grulke et al.31. A number of issues were addressed in this mapping process for the AOP-DB stressor mapping; for example, the same chemical with different chemical identifiers, mapping to the same substance (e.g. Dexamethasone vs Stressor:492 Dexamethasone); spelling errors (e.g. Tacrorimus versus Tacrolimus); general naming that prevents specific mapping to a DTXSID, for instance, “various hydrocarbons”, that is in no way definitive for a particular substance and cannot be mapped to a DTXSID. It should however be noted that ambiguous substances can map to individual chemical structures, to mixtures or to UVCB chemicals (Unknown or Variable Composition, Complex Reaction Products and Biological Materials), for example, as discussed in Williams et al.30. The AOP-DB v.2 stressor linkages are provided through the Dashboard (https://comptox.epa.gov/dashboard/chemical_lists/AOPSTRESSORS), and map EPA chemical substance records to the most current list of AOP stressors (last updated 06/30/21).
AOP-DB Tissue-gene networks
The AOP construct organizes biological and related data from molecular gene targets, both cellular and organ specific, to individual and population levels, so that we can better understand the progression of initiating events of toxicological concern to their benign or adverse outcomes.
The primary aim of the AOP-DB is to organize publicly available data so that users can understand and characterize the biological context of any given AOP. The AOP-DB v.2 addresses the need to understand tissue specific events in an AOP-tissue specific context. Here we implement HumanBase21 data, which includes tissue-specific gene networks for 144 human tissues. By combining genome-wide association studies (GWAS) with tissue-specific gene networks, it is possible to develop tissue-specific gene-interaction networks with probabilities representing the strength of the interaction21. Using the HumanBase API (https://hb.flatironinstitute.org/api/), we import tissue-gene networks for all 144 tissues included in HumanBase and link to each AOP gene. So, for each AOP gene, 144 individual, tissue-gene networks are added. The number of networks will increase as the number of AOP genes in the AOP-DB increases. The final tabular form of the tissue-gene data contains edge data (Entrez1 and Entrez2) representing gene interactions, with individual edge probability scores. Each edge record has a tissue and query gene field, indicating network relationship.
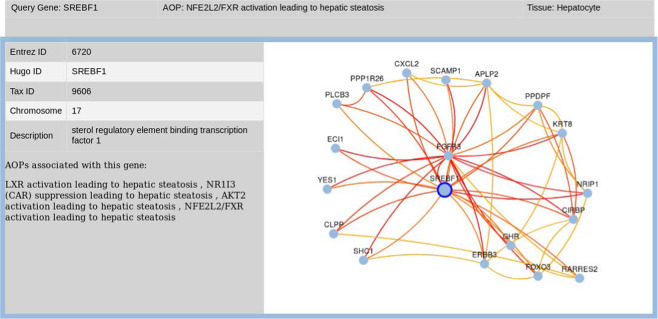
The AOP-DB v.2 implements the HumanBase tissue-gene networks in the creation of a tissue-gene visualizer in the AOP-DB web user interface (Fig. 2). This allows users to navigate to AOP relevant genes and tissue-specific results in a real-time network viewer. The tissue-gene network results can be explored for hypothesis generation and, for example, to understand how tissue-specific AOP activity relates to known disease states in the AOP-DB.
Fig. 2.
AOP Tissue Network Visualization Tool illustrates the tissue-gene network built with user query for SREBF1 for hepatocyte tissue. Associated AOPs for SREBF1 are listed in the left-hand pane.
AOP Haplotypes from 1000 genomes single nucleotide polymorphisms (SNPs)
To our knowledge the AOP-DB v.2 is the first tool that incorporates the novel integration of human population level data with AOP information. AOP-DB v.2 incorporates individual and population level data using data from Ensembl22,23, ENCODE GWAS24, GTex25–27, and the 1000 Genomes Project28,32, with the goal of identifying functionally important AOP-gene variants. The organization of such functional variants for described AOPs makes it possible to identify potential differences in human susceptibility to adverse outcomes on both an individual and group or population level. AOP-DB v.2 incorporates population level SNP frequencies for AOP-genes for five of the geographic super-populations reported by the 1000 Genomes Consortium (European, S. Asian, E. Asian, African, American32) for the Phase 3 data.
In addition, we identified SNPs as functionally important from the ENCODE GWAS catalog24 and the GTEx portal25,26 for each AOP-gene. The SNPs were filtered secondarily using the Ensembl Regulatory Build23 to be located in gene regulatory regions. The resulting SNPs were used to request genotypes using the Ensembl REST API’s variation endpoint for each SNP for each AOP-gene. With these data we can construct individual AOP-gene haplotypes. These data are stored in the Haplotype table of AOP-DB v.2. The resulting functional AOP-gene list for each AOP was then screened using 1000 genomes samples to identify population frequencies of functionally relevant AOP gene SNPs and AOP haplotypes. Figure 3 illustrates the minor allele frequencies (MAF) observed for 104 functionally relevant Human AOP-gene SNPs. These multi-allelic, functional AOP-gene haplotypes can be used for further inquiries to determine levels of variation and significant differences in outcome across population groups.
Fig. 3.
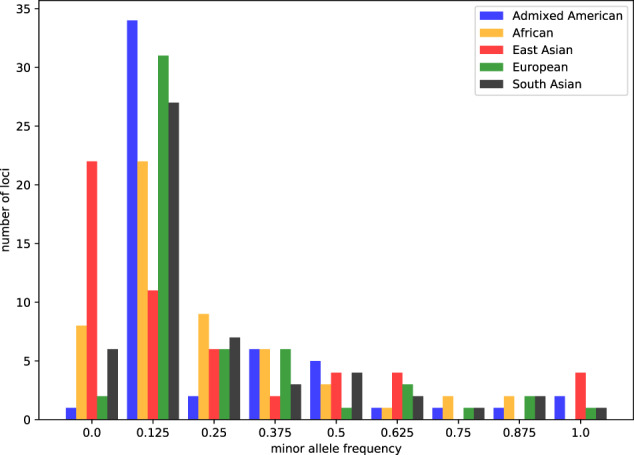
Minor allele frequency (MAF) distribution for SNPs associated with 104 functionally relevant Human AOP-genes for five 1000 Genomes Super populations: American (Blue); African (Yellow); East Asian (Red); European (Green); South Asian (Black).
AOP-DB web-based user interface
The EPA AOP-DB user interface is delivered via a graphical web user interface (coded in Java and React), an application programming interface (API; coded in node.js) and a relational database (MySQL and MariaDB). The AOP-DB web user interface, publicly available at https://aopdb.epa.gov/, has been developed with the goal of providing both data accessibility and data visualization according to the FAIR Principles33 2019b. Basic search functionality of the AOP-DB web user interface allows users who prefer to interact with the web user interface to easily navigate, organize and download AOP and related data to their desktop.
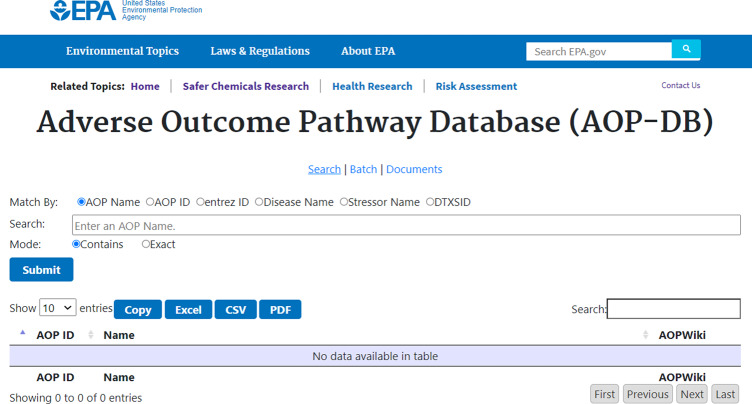
Basic functionality includes a search interface (Fig. 4), batch query tool, and downloads page. The AOP-DB system has been modified for EPA standard visualization as well as integration with current Office of Science and Information Management (OSIM) security protocols for EPA internet hosting. At present, data updates to the AOP-DB occur approximately every 6 months. Initial AOP-DB data visualization has been optimized and includes the following queries: AOP-gene, AOP-disease, AOP-pathway, and AOP-chemical. Search box functionality has also been extended to include autofill processing for inputs. Download capabilities have been extended to include user column select and scroll features, including data download in .csv and .xml, or other useful formats. The ‘Documents’ page links to the AOP-DB v.2 User Manual, which describes all current search functionality enabled for the user interface in detail, and includes sections on maintenance of new records and updates, how content is selected, query tips, and frequently asked questions.
Fig. 4.
AOP-DB search page illustrating “Basic Search” task bar with accepted user input types and download file specificity.
Research description framework mapping of the AOP-DB data tables
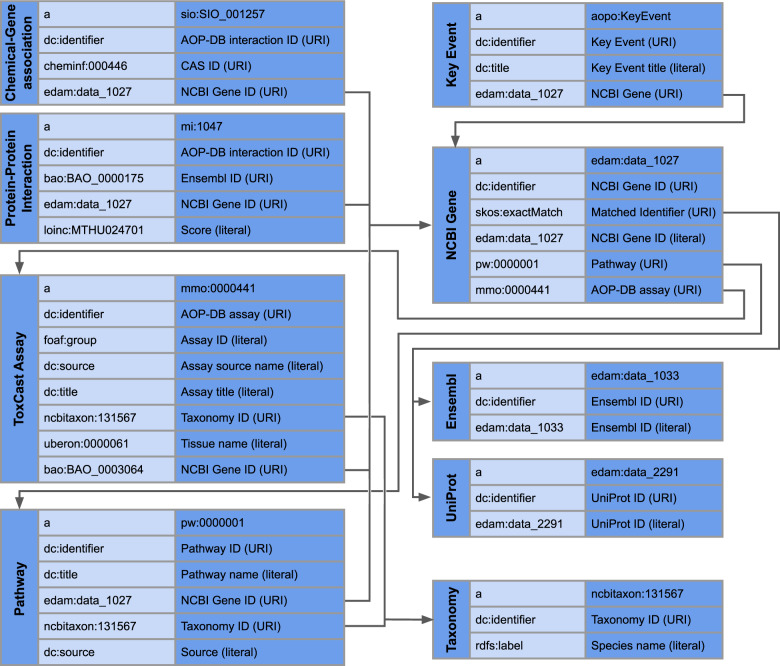
Semantic mapping using a Research Description Framework (RDF)34 was performed for four AOP-DB tables35 as part of an ongoing, international collaboration between the US EPA, OpenRiskNet (https://openrisknet.org/) and researchers at the Department of Bioinformatics – BiGCaT, Maastricht University (http://www.bigcat.unimaas.nl/). Figure 5 illustrates the AOP-DB RDF tables for Chemical-Gene Interaction, Protein-Protein Interaction, ToxCast Assay, and Pathway, and each table’s secondary (and tertiary) keys. A link to the AOP-DB SPARQL endpoint and the workflows generated using these mappings is provided by the OpenRiskNet e-infrastructure (https://openrisknet.org/e-infrastructure/services/147/).
Fig. 5.
AOP-DB Semantic Mapping using the Resource Description Framework (RDF) illustrating Chemical-Gene Interaction, Protein-Protein Interaction, ToxCast Assay, and Pathway tables.
Data Records
Online-only Table 1 describes all the individual data source integrated in the AOP-DB. Data sources are organized according to their biological category, with a short description of the data, along with any manipulations performed, and the URL. The AOP-DB v.2 data frame with all corresponding table data and custom code has been uploaded to a single collection entitled “The Adverse Outcome Pathway Database (AOP-DB) version 2.0”36. The files contained in this collection include the most recent SQL data structure for the AOP-DB v.2, all custom code, tables, and corresponding data categories and keys to create the backend of the database.
Technical Validation
The main quality objective for AOP-DB is ensuring that data included in the data set accurately reflects the data obtained from the raw data source. Data sets considered for curation in the AOP-DB are limited to publicly available, peer-reviewed datasets that have been subjected to rigorous quality control and peer review in their own right. Quality assurance measures to validate the data have focused on ensuring that data records were transcribed accurately from the original data source and represented appropriately in the repository. As a research product of the United States Environmental Protection Agency Office of Research and Development (ORD), the AOP-DB has a Quality Assurance Project Plan (QAPP) which describes each individual data source integrated in the database, how it is implemented, stored and modified. QAPPs describe the necessary quality assurance and quality control measures needed to produce results that meet stated performance criteria. ORD QAPPs are peer-reviewed, approved by management, overseen by a quality assurance manager, and subject to periodic QA and performance quality checks. Figure 1 illustrates, for each data source, each step in obtaining the data from the third-party source, any subroutine that has been created to modify the data, and including any stored local files. In all cases, custom scripts were written to identify and correct any errors in the process of migrating data from source formats into the MySQL database (e.g. duplicated entries).
With public deployment of the AOP-DB v.2, data updates occur approximately every 6 months. Additional, periodic updates will be made to the production version of the database when substantial changes have been made requiring a version update. Changes to the development version of the database will be ongoing and as new, and relevant data become available.
Usage Notes
Though data included in AOP-DB v.2 can be used in many ways in future analyses, users should be aware of limitations of the data set, and appropriate usage of the data. The AOP-DB v.2 incorporates only those AOPs that have molecular identifiers. Many AOPs found in the AOP-Wiki 2.0 do not associate with gene or protein molecular identifiers. This may be due to the nature of the biology described or the state of the science for a particular research area. Users should also be aware that AOP data obtained from the AOP-DB v.2 may be under various levels of review (e.g. Status or SAAOP Status “Do not cite”, or “Included in OECD Workplan”, for example) or the AOP may simply be incomplete. This is due to how AOPs are entered into the AOP-Wiki 2.0 at this time. Additionally, because an AOP may be developed with reference to a particular species, the AOP-DB v.2 reports the gene/protein accession for that species and maps to orthologous gene pairs in humans or other species orthologs when possible, without indication of relevance.
Supplementary information
Acknowledgements
This research contributes to the following FY20–22 research areas of the EPA National Program in Chemical Safety and Sustainability (CSS): Informatics, Synthesis, and Integration (ISI), O-CSS-19, 19.1: Adverse outcome pathway networks; O-CSS 21, 21.3: Computational modeling approaches and case study validation to identify genetic-based susceptibility factors to environmental chemical exposures, 21.7: Quantitative AOP network development for human liver cancer; O-CSS 23.1: Approaches for aligning chemical inventories with AOPs /AOP-based chemical grouping. This research was supported in part by the Research Participation Program at the Office of Research and Development, US Environmental Protection Agency, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between US Department of Energy and US Environmental Protection Agency. This research was additionally funded through a competitive grant for contract support through the Environmental Modeling and Visualization Laboratory. Special thanks to Heidi Paulsen, Tommy Cathey and Tom Transue for their contribution to this effort. Semantic mapping efforts were supported by the European Union funded OpenRiskNet Project. Special thanks to Drs. Marvin Martens, Egon Willighagen, Chris Evelo, and Thomas Exner for their contribution to AOP-DB sematic mapping and training of T.L. The information in this document has been funded wholly or in part by the US Environmental Protection Agency. It does not signify that the contents necessarily reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use. The paper has been subjected to the Agency’s review process and approved for publication. EPA Disclaimer: This manuscript has been reviewed by the Center for Public Health and Environmental Assessment, United States Environmental Protection Agency and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the Agency nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Online Only Table
Author contributions
H.M.M. conceived the project, conceptualized the integration of the existing data sets into the AOP-DB, drafted and edited the manuscript, and led integration of the AOP-DB data with AOP-DB user interface development, OpenRiskNet and the EPA CompTox Chemicals Dashboard. J.S. wrote and maintained scripts to download and extract raw data, maintained underlying MySQL database, improved RDF mapping of AOP-DB tables, and generated tables and figures. T.L. wrote and maintained scripts to download and extract raw data, maintained underlying MySQL database, curated chemicals, and performed the initial RDF mapping of AOP-DB tables. P.L. wrote initial node.js and react code and queries to develop AOP-DB user interface. A.J.W. leads development of the EPA CompTox Chemicals Dashboard and DSSTox registration process for stressor associated chemicals from the AOP-DB.
Code availability
All custom code created to process of manipulate external datasets in the construction or subsequent update of the AOP-DB v.2 relational database tables are made publicly available by the U.S. Environmental Protection Agency, Office of Research and Development (ORD)36.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-021-00962-3.
References
- 1.US Public Law 114–182. Frank R. Lautenberg Chemical Safety for the 21st Century Act (ed 114th Congress, 2016).
- 2.Pittman ME, Edwards SW, Ives C, Mortensen HM. AOP-DB: A database resource for the exploration of Adverse Outcome Pathways through integrated association networks. Toxicol Appl Pharmacol. 2018;343:71–83. doi: 10.1016/j.taap.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortensen HM, et al. Leveraging human genetic and adverse outcome pathway (AOP) data to inform susceptibility in human health risk assessment. Mamm Genome. 2018;29:190–204. doi: 10.1007/s00335-018-9738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patlewicz G, Worth AP, Ball N. Validation of Computational Methods. Adv Exp Med Biol. 2016;856:165–187. doi: 10.1007/978-3-319-33826-2_6. [DOI] [PubMed] [Google Scholar]
- 5.Delrue N, et al. The adverse outcome pathway concept: A basis for developing regulatory decision-making tools. Altern Lab Anim. 2016;44:417–429. doi: 10.1177/026119291604400504. [DOI] [PubMed] [Google Scholar]
- 6.Sakuratani Y, Horie M, Leinala E. Integrated Approaches to Testing and Assessment: OECD Activities on the Development and Use of Adverse Outcome Pathways and Case Studies. Basic Clin Pharmacol Toxicol. 2018;123(Suppl 5):20–28. doi: 10.1111/bcpt.12955. [DOI] [PubMed] [Google Scholar]
- 7.Ives C, Campia I, Wang RL, Wittwehr C, Edwards S. Creating a Structured AOP Knowledgebase via Ontology-Based Annotations. Appl In Vitro Toxicol. 2017;3:298–311. doi: 10.1089/aivt.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis AP, et al. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2017;45:D972–D978. doi: 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamburov A, et al. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jassal B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinero J, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geer LY, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.US EPA. ToxCast Assay Target mappings from invitroDBv3.2 database. https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data (2017).
- 18.Dix DJ, et al. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci. 2007;95:5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- 19.Judson RS, et al. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect. 2010;118:485–492. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kavlock R, et al. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol. 2012;25:1287–1302. doi: 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- 21.Greene CS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt, S. E. et al. Ensembl variation resources. Database (Oxford) 2018 10.1093/database/bay119 (2018). [DOI] [PMC free article] [PubMed]
- 23.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GTEx Consortium The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GTEx Consortium Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017;49:1664–1670. doi: 10.1038/ng.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.1000 Genomes Project An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard AM, Williams CR. Distributed structure-searchable toxicity (DSSTox) public database network: a proposal. Mutat Res. 2002;499:27–52. doi: 10.1016/s0027-5107(01)00289-5. [DOI] [PubMed] [Google Scholar]
- 30.Williams AJ, et al. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. J Cheminform. 2017;9:61. doi: 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grulke, C. M., A. J. Williams, Thillanadarajah, I., Richard, A. M. EPA’s DSSTox database: History of development of a curated chemistry resource supporting computational toxicology research. Computational Toxicology12 (2019). [DOI] [PMC free article] [PubMed]
- 32.1000 Genomes Project A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson MD, et al. Evaluating FAIR maturity through a scalable, automated, community-governed framework. Sci Data. 2019;6:174. doi: 10.1038/s41597-019-0184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.RDF Working Group. https://www.w3.org/RDF/ (2014).
- 35.Mortensen, H. M.; Levey, T.A.; Martens, M.; Evelo, C.T; Willighagen, E. L. Enhancing the EPA Adverse Outcome Pathway Database (AOP-DB): Recent Updates and Sematic Integration Toxicological Sciences174 (2020).
- 36.Mortensen HM, 2021. The EPA Adverse Outcome Pathway Database version 2.0 (AOP-DB_v2) U.S. EPA Office of Research and Development (ORD) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Mortensen HM, 2021. The EPA Adverse Outcome Pathway Database version 2.0 (AOP-DB_v2) U.S. EPA Office of Research and Development (ORD) [DOI]
Supplementary Materials
Data Availability Statement
All custom code created to process of manipulate external datasets in the construction or subsequent update of the AOP-DB v.2 relational database tables are made publicly available by the U.S. Environmental Protection Agency, Office of Research and Development (ORD)36.