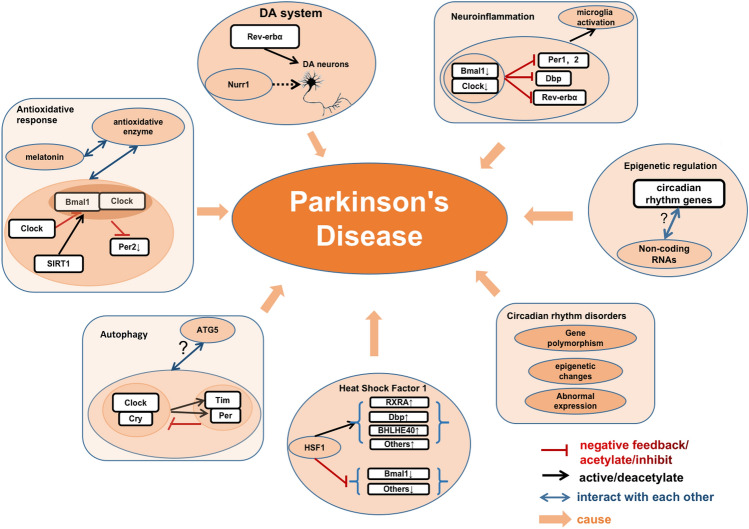
Fig. 2.
Circadian rhythm genes and Parkinson’s disease. Circadian rhythm genes participate in PD pathology via various pathways. The Clock/Cry complex activates Tim and Per expression to form complexes and inhibits Clock/Cry complex activity. This network interacts with ATG5 to regulate autophagy in PD. Clock acetylates Bmal1, and inhibits the Bmal1/Clock-dependent transcription of Per2. SIRT1 deacetylates Per2 and Bmal1. This network along with melatonin interacts with antioxidant enzymes yielding an antioxidative response in PD. Bmal1 and Clock are lower in PD and the Bmal1/Clock complex-dependent transcription of Per1, Per2, Dbp, and Rev-erbα is inhibited. This network activates microglia-dependent neuroinflammation in PD. Rev-erbα competitively cooperates with Nurr1 to control DA gene transcription and the development and function of DA neurons. Some HSF1 targets that are associate with circadian rhythm genes (RXRA, Bhlhe40, and DBP) are upregulated, while some others (Bmal1) are inhibited. Non-coding RNAs undertake epigenetic regulation of the circadian rhythm system, but the mechanism is not clear. Circadian rhythm alterations, including abnormal expression of circadian rhythm genes, epigenetic changes, and gene polymorphisms, participate in the pathogenesis of PD.