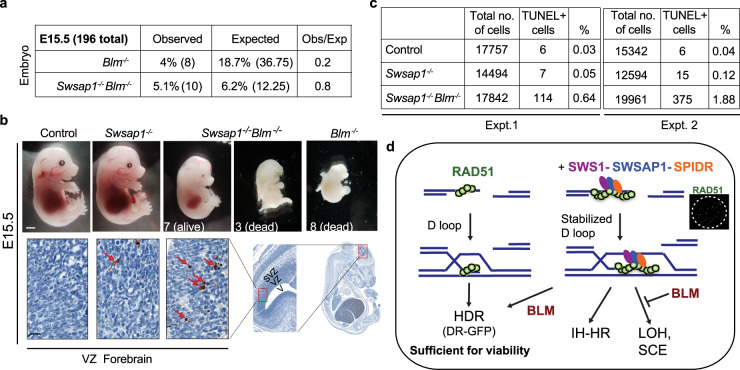
Fig. 4. Loss of SWSAP1 prolongs embryonic survival of Blm mutants.
a Blm−/− embryos at E15.5 are recovered at five-fold lower than expected from the Mendelian ratio, however, Swsap1−/− Blm−/− embryos are recovered only 20% less often than expected. (See also Supplementary Fig. 9f). b Analysis of brains from E15.5 embryos. Although Swsap1−/− embryos have normal embryonic development, the few Blm−/− embryos recovered at this stage are dead. By contrast, most Swsap1−/− Blm−/− are alive, but smaller in size, although a few resemble Blm−/− embryos. Scale bar, 1 mm. Few apoptotic cells are observed in the ventricular zone (VZ) of the forebrain of Swsap1−/− embryos, while Swsap1−/− Blm−/− embryos show numerous TUNEL-positive cells (red arrows). The region analyzed in embryos is progressively depicted (red rectangles), with the ventricle (V), ventricular zone (VZ), and sub-ventricular zone (SVZ) indicated. Scale bar, 50 μm. (See also Supplementary Fig. 10a, b). c Quantification of TUNEL positive cells in the VZ from two experiments. d Model for the role of the SWS1–SWSAP1–SPIDR complex and its genetic interaction with BLM in multiple HDR outcomes. Right, SWS1–SWSAP1–SPIDR functions to stabilize RAD51 nucleoprotein filaments, which can give rise to visible foci and stabilized D loops. These stabilized intermediates are required for IH-HR and the efficient formation of dHJs. dHJs can be dissolved by BLM or resolved by strand nicking to generate crossovers with associated LOH when homologs are involved or SCEs when sister chromatids are involved. Left, smaller, or less stable RAD51 filaments in the absence of SWS1–SWSAP1–SPIDR are sufficient for intrachromosomal HDR through the synthesis-dependent strand annealing pathway, for example, as assayed in the DR-GFP reporter. In addition to dissolution, BLM is known to unwind D loops to promote synthesis-dependent strand annealing; the requirement for this activity may be less in SWS1–SWSAP1–SPIDR mutant cells since RAD51 nucleoprotein filaments are predicted to be inherently less stable, partially restoring HDR, as assayed with the DR-GFP reporter or by gene targeting.