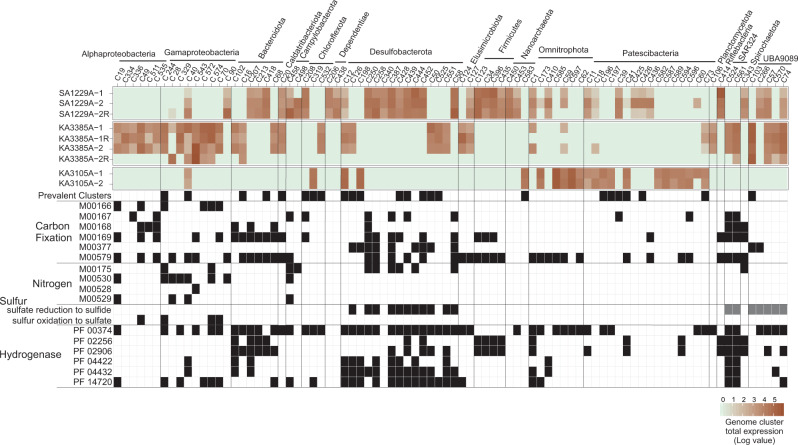
Fig. 5. Expression profile and metabolic context of the highly expressing genomic clusters.
The expression profile of genome clusters with a total expression ≥ 10,000 TPM and their metabolic potential for nitrogen and sulfur energy metabolism as well as carbon fixation is represented as KEGG modules presence only if all genes of the module or the key genes of the process are present (in black). Reductive pentose phosphate cycle, ribulose-5P ≥ glyceraldehyde-3P (M00166), reductive pentose phosphate cycle, glyceraldehyde-3P ≥ ribulose-5P (M00167), Crassulacean acid metabolism, dark (M00168) and light (M00169), reductive acetyl-CoA pathway (Wood-Ljungdahl pathway) (M00377), and phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA ≥ acetate (M00579). Nitrogen fixation (M00175), dissimilatory nitrate reduction (M00530), nitrification (M00528), and denitrification (M00529). Genes involved in the dissimilatory sulfur metabolism are shown in Supplementary Data 7. Squares highlighted in gray contain genes for sulfate reduction to sulfide but are missing dsrD. Nickel-dependent hydrogenase (PF00374), iron hydrogenase (PF02256 and PF02906), coenzyme F420 hydrogenase/dehydrogenase (PF04422 and PF04432), and NiFe/NiFeSe hydrogenase (PF14720).