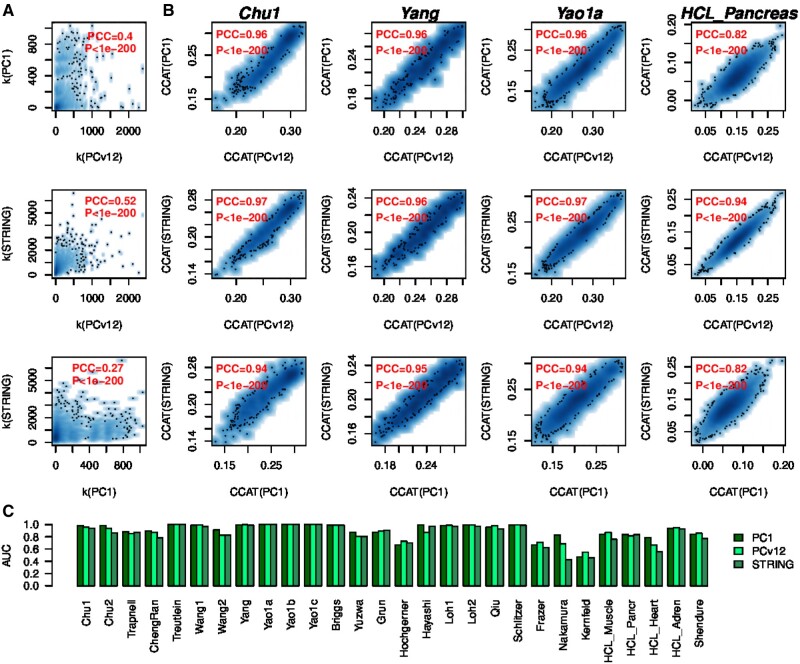
Fig. 3.
Robustness of CCAT to choice of PPI network. (A) Scatterplots of the degree of each protein between each pair of PPI networks (PC1 = Pathway Commons V1, PCv12 = Pathway Commons v12, STRING=STRING database v11). The Pearson Correlation Coefficient (PCC) and associated P-value are shown in red. (B) Scatterplots of the CCAT estimates derived from each of the three PPI networks and in each of four independent scRNA-Seq studies (Chu1, Yang, Yao1a and HCL-Pancreas). The Pearson Correlation Coefficient (PCC) and associated P-value are shown in red. (C) Barplots displaying the accuracy (AUC) of CCAT to discriminate high and low potency cells as a function of PPI network used and scRNA-Seq dataset