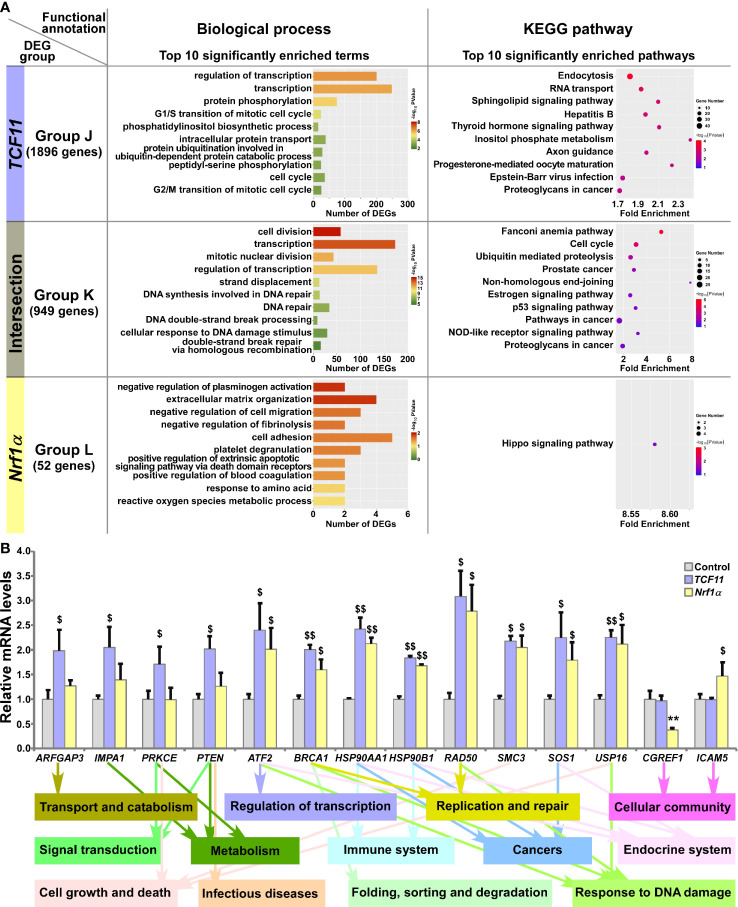
Figure 6.
Functional annotation of specific or common DEGs in TCF11 and Nrf1α cells. (A) Top 10 of significant biological process terms and pathways enriched by DEGs in Groups J, K, and L were exhibited in histograms and scatterplots, respectively. (B) After induced with 1 μg/ml Tet for 12 h, total RNAs were isolated from Control, TCF11 or Nrf1α cell lines and then reversely transcribed into the first strand of cDNA. Subsequently, relevant mRNA levels of DEGs that were associated with more functions as annotated, along with high expression levels and well significance in Groups J to L, were determined by quantitative real-time PCR in Control, TCF11 and Nrf1α cells. The data are shown as mean ± SEM (n = 3 × 3, **p <0.01; $ p <0.05; $$ p <0.01, when compared to the Control values).