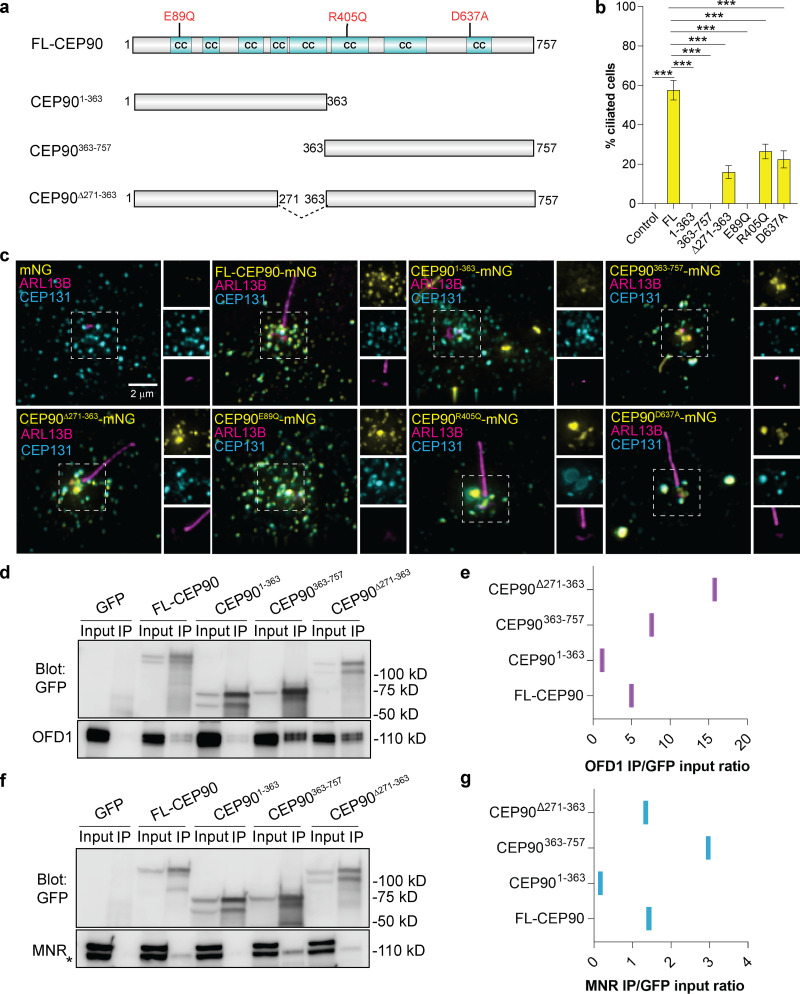
Figure 5.
CEP90 ciliopathy mutations affect ciliogenesis and centriolar satellite morphology. (a) Schematic representation of full-length (FL) human CEP90, truncation constructs, and disease-associated mutations. Coiled-coil (CC) domains identified by MARCOIL (Zimmermann et al., 2018) at 90% threshold are indicated in cyan. (b) Quantification of ciliation frequency of CEP90−/− RPE1 (control) and CEP90−/− cells expressing mNeonGreen-tagged disease variants and truncations of CEP90. Graph shows mean ± SEM. ***, P < 0.05, one-way ANOVA. n > 100 cells from two biological replicates. (c) CEP90−/− RPE1 cells and CEP90−/− cells expressing the denoted mNeonGreen (mNG)–tagged versions of CEP90 were serum starved for 24 h and immunostained for cilia (ARL13B), centriolar satellites (CEP131), and mNeonGreen. Scale bar = 2 µm. (d) Immunoprecipitation of GFP-tagged full-length and truncation constructs of CEP90 blotted for GFP and OFD1. (e) Quantification of OFD1 band intensities relative to corresponding GFP input band intensities. (f) Immunoprecipitation of GFP-tagged full-length and truncation constructs of CEP90 blotted for GFP and MNR. (g) Quantification of MNR band intensities relative to corresponding GFP input band intensities. Asterisk represents the MNR band; the top band is nonspecific.