Figure 5. Vessel Topography Drives Cell Residence Time and Occlusion during Initial Dissemination through the Circulatory System.
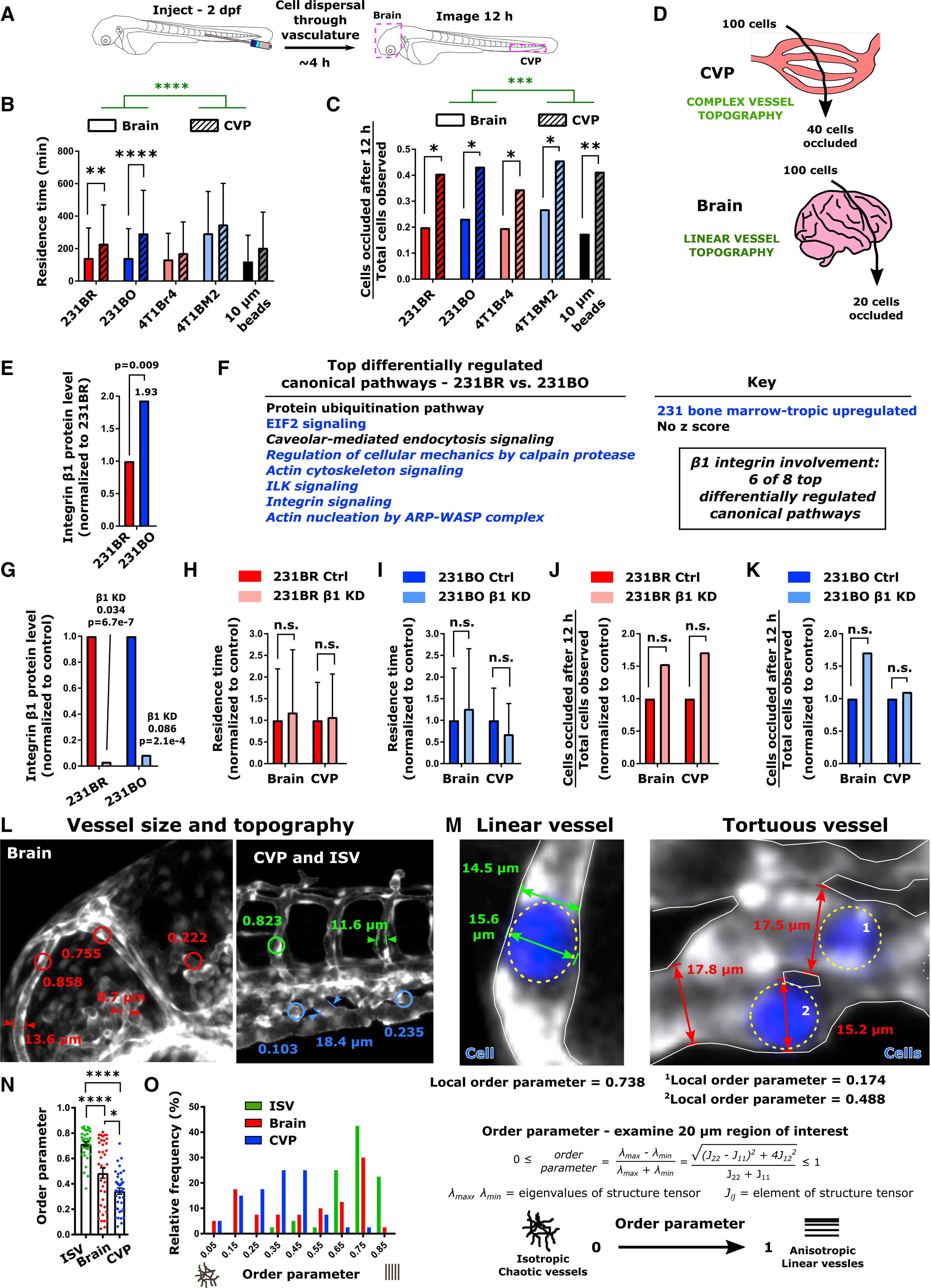
(A) Schematic of experiment. 2 days post-fertilization (dpf) zebrafish were injected with MDA-MB-231 brain-tropic (231BR) or bone-marrow-tropic (231BO) cells, 4T1 brain-tropic (4T1Br4) or bone-marrow-tropic (4T1BM2) cells, or 10-μm-diameter polystyrene beads (beads) in the posterior cardinal vein. Following dispersal through the vasculature, the brain and CVP of each larva were imaged for 12 h via confocal microscopy.
(B) Residence time (mean ± SD) of cells and beads through the brain and CVP. Averages were calculated from the pool of all cells or beads imaged within the indicated organ. Tissue of observation (****p < 0.0001, F = 25.36, DF = 1) and particle/cell type (p < 0.0001, F = 13.65, DF = 4) were significant sources of variation by two-way ANOVA. Additionally, residence times of 231BR and 231BO cells were significantly less in the brain than the CVP (**p = 0.0012; ****p < 0.0001 by Sidak’s multiple comparisons test).
(C) Fraction of cells or beads occluded in the brain and CVP at the end of the 12 h imaging period from the total number of particles observed in each organ over the imaging period. The tissue of observation but not particle/cell type was a significant source of variation by two-way ANOVA (***p = 0.0002, F = 184.2, DF = 1), and occlusion fractions were significantly higher in the CVP than the brain for all particles tested by Sidak’s multiple comparisons test (231BR, *p = 0.0156; 231BO, *p = 0.0172; 4T1Br4, *p = 0.0486; 4T1BM2, *p = 0.0217; beads, **p = 0.0090).
(D) Schematic of cell occlusion in the brain and CVP. Regardless of cell type tested, ~40% of cells observed became occluded in the CVP and ~20% of cells observed became occluded in the brain over the 12 h imaging period tracking initial cell dissemination. For (B) and (C), values were calculated from ensemble data across 7 larvae for 231BR cells (N = 140 cells in the brain, N = 170 cells in the CVP), 10 larvae for 231BO cells (N = 86 cells in the brain, N = 97 cells in the CVP), 6 larvae for 4T1Br4 cells (N = 61 cells in the brain, N = 78 cells in the CVP), 8 larvae for 4T1BM2 cells (N = 26 cells in the brain, N = 70 cells in the CVP), and 6 larvae for polystyrene beads (N = 183 beads in the brain, N = 29 beads in the CVP).
(E) Relative protein expression of integrin β1 in MDA-MB-231 organ-tropic cell lines from mass spectrometry data. Expression in bone-marrow-tropic 231BO cells is normalized to that in brain-tropic 231BR cells. Fold changes in protein level and p values are indicated.
(F) List of the top differentially regulated canonical signaling pathways between MDA-MB-231 brain-tropic (231BR) and bone-marrow-tropic (231BO) cells from pathway analysis of mass spectrometry data. Where possible, the direction of pathway regulation (from IPA Analysis Z score) is indicated (blue, upregulated in 231BO; black, no conclusive direction of regulation). Pathways in which β1 integrin are implicated are italicized.
(G) Knockdown of integrin β1 assessed from mass spectrometry data. β1 integrin levels are normalized within each cell line relative to treatment with control siRNA. Fold changes and p values are indicated.
(H and I) Residence time (average ± SD) of (H) 231BR cells and (I) 231BO cells in the brain and CVP upon β1 integrin silencing, normalized to the average residence time in the same organ for control cells. Integrin β1 silencing was not a significant source of variation by two-way ANOVA (231BR: p=0.4679; 231BO: p=0.8425), and differences between control and knockdown cells in a given organ were not significant (n.s.) by Sidak’s multiple comparisons test (231BR: p=0.7247 in brain, p=0.9413 in CVP; 231BO: p=0.3234 in brain, p=0.1676 in CVP).
(J and K) Fraction of (J) 231BR or (K) 231BO β1 integrin knockdown cells passing through a given organ that became occluded over the 12 h imaging period tracking initial cell dissemination, normalized to the fraction in the same organ for control cells. Fractions were calculated from the pool of all cells imaged in the brain and CVP for a given condition. Integrin β1 silencing was not a significant source of variation by two-way ANOVA (231BR: p=0.0925; 231BO: p=0.4067), and differences between control and knockdown cells in a given organ were not significant (n.s.) by Sidak’s multiple comparisons test (231BR: p=0.1766 in brain and CVP; 231BO: p=0.6480 in brain and CVP). For (H–K), residence times and occlusion fractions were calculated from: 231BR Ctrl – 3 larvae, N = 30 cells in brain, N = 35 cells in CVP; 231BR integrin β1 knockdown – 4 Larvae, N = 94 cells in brain, N = 74 cells in CVP; 231BO Ctrl – 3 larvae, N = 78 cells in brain, N = 70 cells in CVP; 231BO integrin β1 knockdown – 4 larvae, N = 43 cells in head, N = 55 cells in CVP.
(L) Representative vessel width and order parameter measurements in the intersegmental vessels (ISV), Caudal Vein Plexus (CVP), and brain of 2 days post-fertilization (dpf) Tg(fli1:EGFP) zebrafish. Order parameters were measured over 20-μm-diameter circular regions of interest.
(M) Representative images of linear and tortuous zebrafish blood vessels. Cancer cells are shown in blue and outlined by dashed yellow ovals. Representative examples of cell and vessel width measurements are illustrated. Additionally, the local order parameter describing vessel topography as defined is given in the vicinity of each indicated cell.
(N) Average ± SD and (O) distribution of the order parameter characterizing the topography of vessels in the ISV, brain, and CVP over 20-μm-diameter circular regions of interest. A value of 1 indicates aligned structures, while a value of 0 indicates isotropic structures. Vessels were measured from four Tg(fli1:EGFP) zebrafish at 2 dpf, 10 regions per tissue per fish, to obtain a total of N = 40 vessels at each location. *p < 0.05; ****p < 0.0001 by Dunn’s multiple comparisons post-test following Kruskal-Wallis test (p < 0.0001). See also Figure S5.
