Figure 6|. Type I-B subtype 2 PmcCAST system homes to tRNA-val.
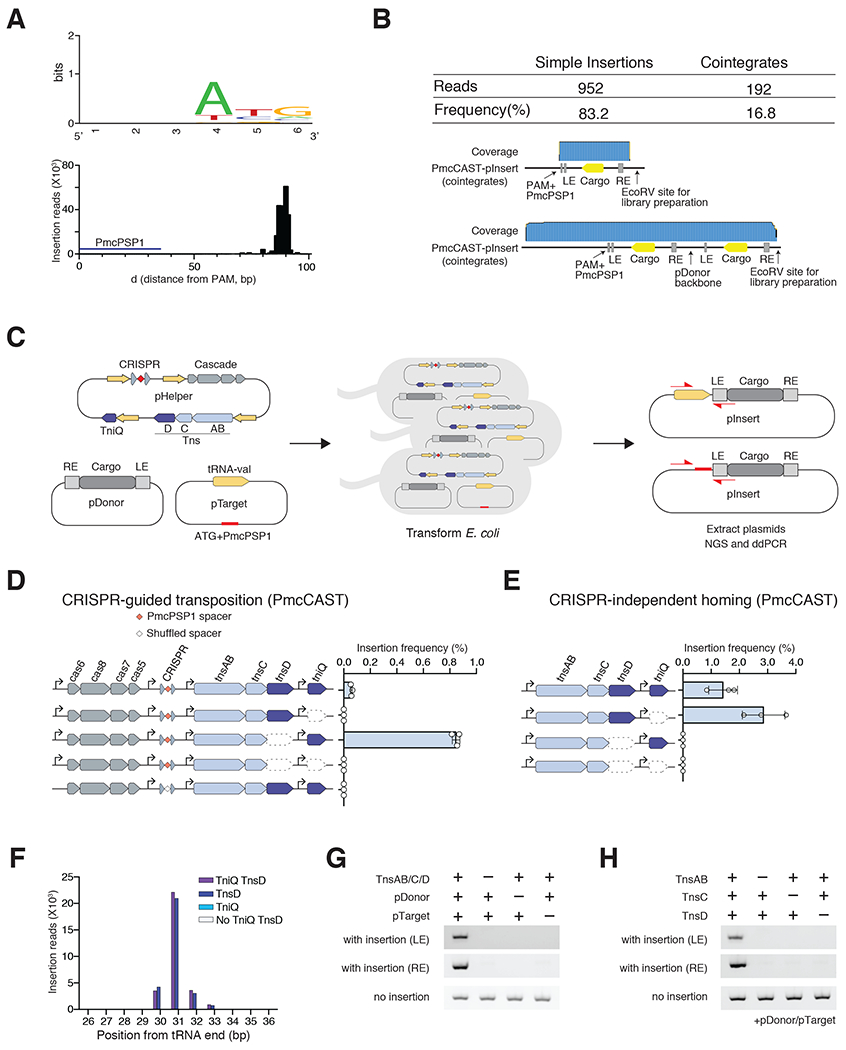
(A) Top. PAMs for PmcCAST RNA-guided insertions. Bottom. AvCAST RNA-guided insertion positions identified by deep sequencing. (B) Long-read Nanopore sequencing to characterize the structure of pInsert. (C) Schematic of experiment to compare RNA-guided transposition and homing to tRNA-val by PmcCAST in E.coli. (D) RNA-guided insertion frequency of PmcCAST into the pTarget with PmcPSP1 and tRNA-val gene at each ΔTniQ or ΔTnsD condition. Data are represented as mean ± SD. (E) Tn7-like machinery-mediated insertion frequency of PmcCAST into pTarget at each ΔTniQ or ΔTnsD condition. Data are represented as mean ± SD. (F) PmcCAST-mediated prominent insertion at tRNA-val gene on target plasmid identified by deep sequencing. Purple bar indicates the position by all Tns proteins (TnsAB, TnsC, TniQ, and TnsD). Blue bar indicates the position at ΔTniQ condition. Light blue bar indicates the position at ΔTnsD condition. White bar indicates the position at ΔTniQΔTnsD conditions. (G) Tn7-like machinery and donor requirements for in vitro transposition on tRNA-val gene. pInsert was detected by PCR for LE and RE junctions. (H) Tns protein requirements for in vitro transposition on tRNA-val gene. All reactions contained pDonor and pTarget.
