Figure 1. Structures of AcrB and homolog solved in detergent-based and detergent-free systems.
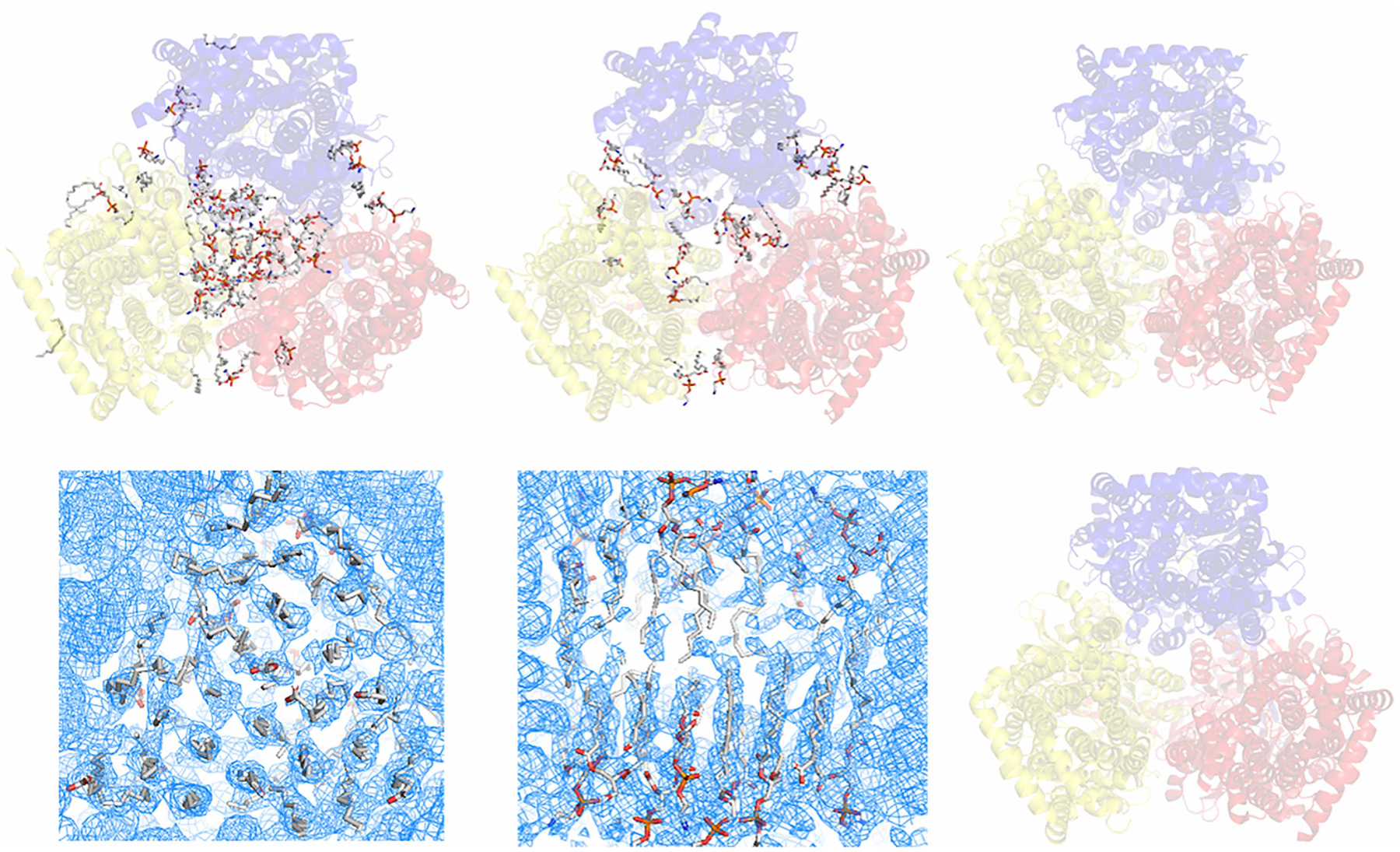
(A) Single-particle cryo-EM structure of E. coli AcrB determined using NCMN system (PDB: 6BAJ). The NCMN system retains native cell membrane lipids associated with AcrB, a native lipid bilayer patch composed of 24 lipid molecules can be unambiguously determined. (B) Single-particle cryo-EM structure of Neisseria gonorrhoeae MtrD (AcrB homolog) determined using Nanodisc technology (PDB: 6VKS). Nanodisc reconstitution using artificial lipids or total E. coli lipids could not reconstruct the native lipid bilayer patch within the lipid cavity of the transmembrane domain. (C) Single-particle cryo-EM structure of AcrB from Salmonella enterica subsp. enterica (PDB: 6Z12) determined using SMALP technology. (D,E) Single-particles cryo-EM density of E. coli AcrB determined using Saposin-lipid reconstitution (EMD:10185). Close-up view of the lipid plug (D) bottom view of the inner leaflet lipid-pattern with a similar orientation to the structure in panel A, but with a clockwise rotation around the Z-axis about 30° for a better view; (E) side view of the lipid bilayer plug, turn 90° clockwisely around the X-axis relative to the density in panel (D); top half: outer leaflet, bottom half: inner leaflet). Saposin-lipid reconstitution can partially reconstitute the lipid bilayer patch within the lipid cavity of the transmembrane domain of E. coli AcrB. However, it still could not recover the native state of the lipid bilayer structure. (F) Crystal structure of E. coli AcrB using detergent-based method. In this structure, almost all native lipids were washed away by detergents. In all the figures above, cartoons show the AcrB protein. Sticks show the lipid molecules.
