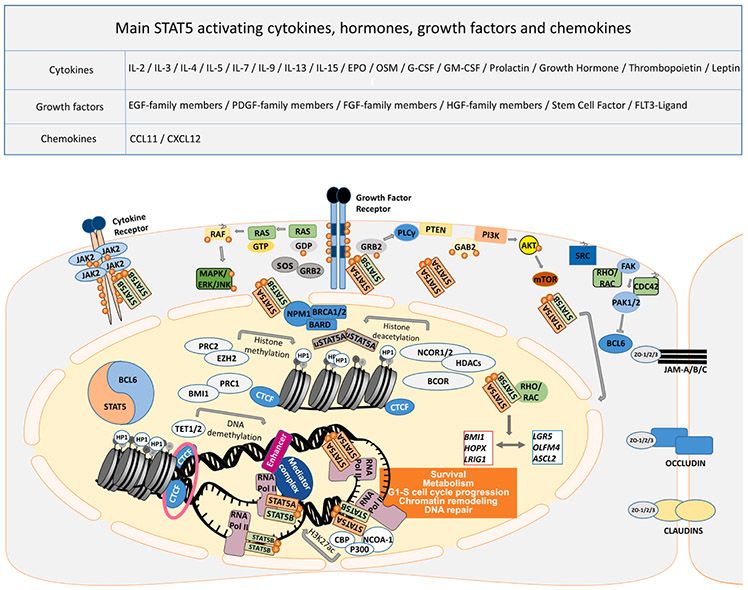
Fig. 1.
Cell signaling pathways involved in cytokine, hormone, growth factor and chemokine signaling in association with STAT5 functions. Signaling pathways, triggered by a plethora of cytokines, hormones, growth factors, and chemokines regulate survival, metabolism, and G1-S cell cycle progression, as well as chromatin remodeling through STAT5 involvement with and without pYSTAT5 function. Overexpression of growth factor/cytokine receptor and/or mutated downstream effectors are often found in the EGFR-RAS-RAF axis and PI3K-AKT-mTOR pathway. In the cytoplasm, STAT5A/B can be activated through phosphorylation of tyrosine residue at position 694/699 by JAK or tyrosine kinase receptor. STAT5A is translocated into the nucleus with the assistance of RHO/RAC, which phosphorylates a serine residue of STAT5A at position 779. There are complex homo- and heterodimers with STAT5B, and RHO/RAC can bind to serine phosphorylated STAT5A homo- or STAT5A/B heterodimers, forming a trimer. Besides, higher-order oligomer formation is facilitated by higher pYSTAT5 levels in a concentration-dependent manner. RHO and RAC can both trigger PAK1/2 kinase activation, which phosphorylates STAT5A to promote nuclear shuttling, or PAK kinases block the BCL6 transcription factor by serine phosphorylation. Interestingly, STAT5 binds to DNA as a transcription factor, competing with BCL6 for identical DNA binding sites, to promote transcription of the target genes. As discussed above, BCL6 acts as a locus repressor of STAT5A/B, autoregulating itself by two unique and distinct promoters. STAT5 can also recruit epigenetic modifiers, such as NCOR1/2, BCOR, HDAC3, EZH2, TET1/2 and CBP-P300. BCL6 and STAT5 also compete for corepressor binding. Even though only NCOR1/2 mapping was reported, BCOR attraction might be relevant as well. Furthermore, STAT5 acts on chromatin topology to consequently regulate the expression of genes, keeping the balance between renewal- (LGR5, OLFM4, ASCL4) and quiescence-associated genes (BMI1, HOPX, LRIG1). Surprisingly, unphosphorylated STAT5A (uSTAT5A) binds to heterochromatin protein 1 (HP1) to promote chromatin compaction. In comparison, the more oncogenic STAT5B gene product binds DNA more abundantly, which is antagonized by STAT5A, being itself under transcriptional regulation by TP53 action. Moreover, uSTAT5 was shown to be colocalized with CTCF in myeloid cells. CTCF transcription factor binding organizes chromatin into topology associated domains, where open and closed chromatin is folded by simplified schematic loop structures in association with Cohesin subunits (pink ring), separating them into different transcriptionally active or closed regions. Cohesin subunits, STAG1/2 are often mutated in colorectal cancer.