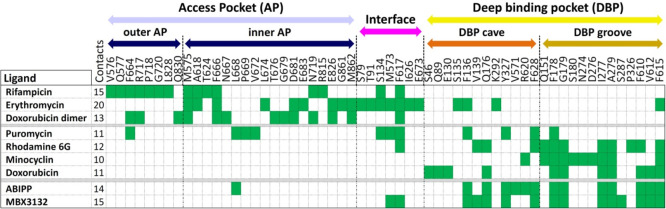
Table 3. Assignment of Potentially Interacting Residues with Substrates Found in Crystal Structuresa.
All residues with atoms found in a radius of 3.5 Å around one of the co-crystallized ligands were assigned according to their localization to the outer AP (cleft), the AP-DBP interface, and the DBP cave and groove regions. Potential interactions with a substrate are indicated in green. The numbers after the ligand/substrate give the sum of potential interacting residues per substrate. Rifampicin and the doxorubicin dimer are found exclusively interacting with residues of the AP and the interface region between the AP and DBP, while erythromycin also interacts with parts of the DBP cave and groove. Puromycin, binding underneath the switch loop, makes contact to all five indicated sections. Rhodamine 6G, minocycline, and doxorubicin bind to the DBP, primarily (but not only) to the groove region. The inhibitors, ABIPP and MBX3132 as a consequence of their orthogonal orientation, are interacting with both groove and cave regions of the DBP. Adapted with permission from Kobylka et al.342 Copyright 2019, Kobylka, Kuth, Müller, Geertsma & Pos under Creative Commons Attribution 4.0 International Public License: https://creativecommons.org/licenses/by/4.0/legalcode.