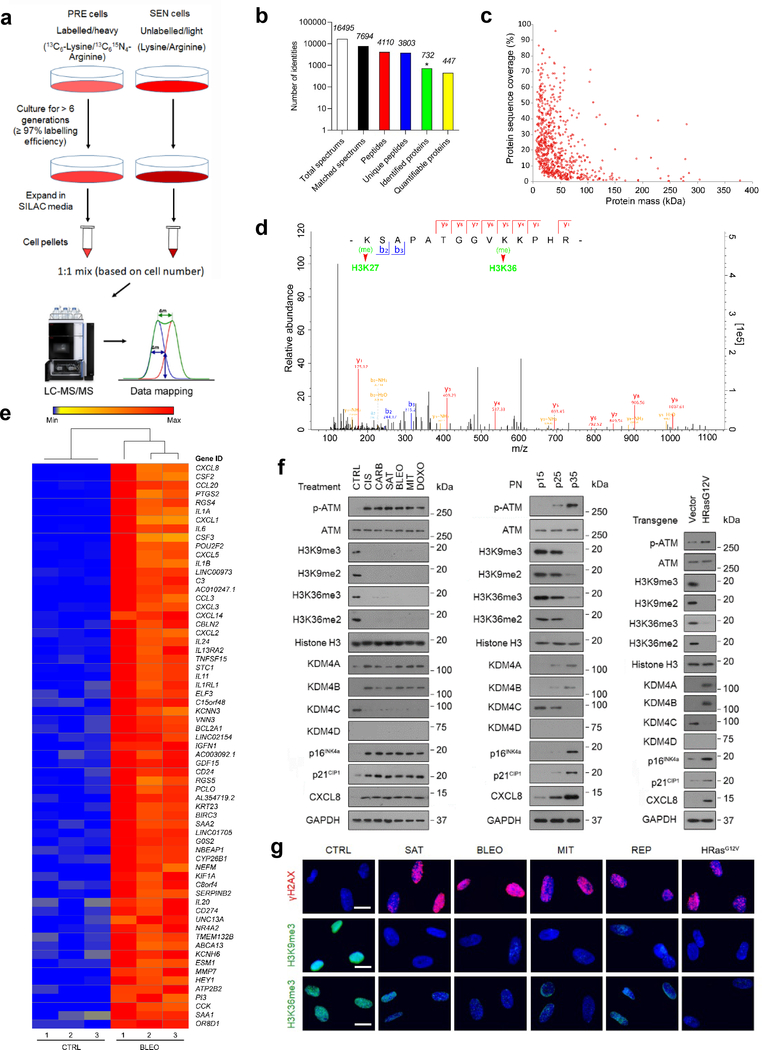
Fig. 1. Histone H3 Lysine Sites Are Epigenetically Modified upon Cellular Senescence.
a, Technical scheme of SILAC-based identification of intracellular proteins in presenescent versus senescent cells of the human stromal line PSC27. PRE cells, presenescent cells. SEN cells, senescent cells. b, Column statistics of different categories of protein molecules in output data after SILAC analysis. *, identified proteins (732). Data representative of 3 independent experiments. c, Scatterplot of proteins identified by SILAC procedure. Protein sequence coverage was plotted against protein mass (447 quantifiable). d, A representative plot derived from characterization of tandem mass spectrometry (MS/MS) based quantitative proteomics profiling. For MS scans, the m/z scan range was 100 to 1100. Intact peptides were detected in the Orbitrap at a resolution of 70,000. e, Heatmap depicting genes significantly upregulated in SEN cells after bleomycin treatment. CTRL, control. BLEO, bleomycin. Genes are ordered by their expression fold change (highest on top) in PRE versus SEN cells after RNA-seq. f, Immunoblot analysis of key molecules in DNA damage repair, cellular senescence and the SASP in PSC27 cells induced to senescence by chemotherapeutic agents (TIS), replicative exhaustion (RS), or oncogene activation (OIS). PN, passage number. p15, p25, p35 representing different passages in culture. Vector, empty control for human HRasG12V. H3K9me2/3 and H3K36me2/3, histone H3 methylation markers. g, Immunofluorescence staining of γH2AX, H3K9me3, and H3K36me3 in PSC27 cells after chemotherapeutic treatment (SAT, BLEO, and MIT), replicative exhaustion (REP), or oncogene activation (HRasG12V). SAT, satraplatin. BLEO, bleomycin. MIT, mitoxantrone. Scale bars, 5 μm. Data in f-g are representative of 3 independent biological replicates.