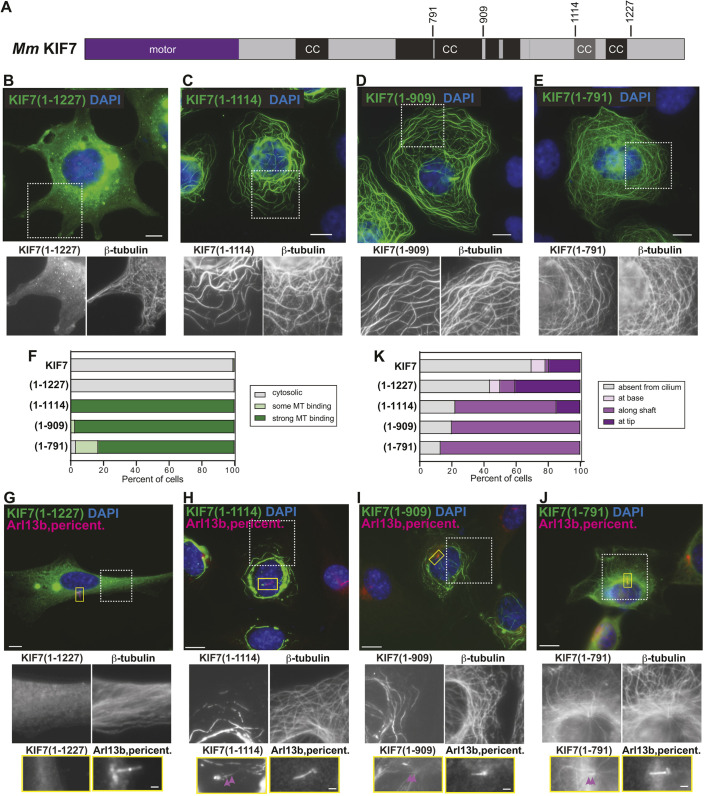
Fig. 2.
Truncations that remove the inhCC are active for microtubule binding. (A) Domain organization of full-length KIF7. The positions of the truncations at amino acids 791, 909, 1114 and 1227 are indicated on top. CC, coiled coil; black box, CC prediction >90%; gray box, CC prediction >80%, according to COILS program. (B-F) COS-7 cells expressing the indicated mCit-tagged truncated versions of KIF7 were fixed and stained with an antibody against β-tubulin to mark microtubules and with DAPI to mark the nucleus. The white boxed region is shown below with separate channels for KIF7-mCit and microtubules. (F) Quantification of microtubule (MT) binding. n=136 (KIF7), 132 [KIF7(1-1227)], 123 [KIF7(1-1114)], 149 [KIF7(1-909)] and 125 [KIF7(1-791)] cells across at least three independent experiments. (G-K) NIH-3T3 cells expressing the indicated mCit-tagged truncated versions of KIF7 were fixed and stained with antibodies against β-tubulin to mark microtubules, Arl13b to mark the primary cilium, pericentrin to mark the basal body and DAPI to mark the nucleus. The white boxed region is shown below with separate channels for KIF7-mCit and microtubules. The primary cilium in the yellow boxed region is shown below with separated images of KIF7 and the cilium markers. Purple arrowheads indicate localization along the cilium shaft. (K) Quantification of cilium localization. n=46 (KIF7), 32 [KIF7(1-1227)], 27 [KIF7(1-1114)], 25 [KIF7(1-909)] and 23 [KIF7(1-791)] cells across at least three independent experiments. Scale bars: 10 µm (B-E,G-J); 1 µm (G-J, bottom panels).