Supplemental Digital Content is available in the text.
Key Words: PD-L1, SP142, triple-negative breast cancer
Abstract
SP142 programmed cell death ligand 1 (PD-L1) status predicts response to atezolizumab in triple-negative breast carcinoma (TNBC). Prevalence of VENTANA PD-L1 (SP142) Assay positivity, concordance with the VENTANA PD-L1 (SP263) Assay and Dako PD-L1 IHC 22C3 pharmDx assay, and association with clinicopathologic features were assessed in 447 TNBCs. SP142 PD-L1 intraobserver and interobserver agreement was investigated in a subset of 60 TNBCs, with scores enriched around the 1% cutoff. The effect of a 1-hour training video on pretraining and posttraining scores was ascertained. At a 1% cutoff, 34.2% of tumors were SP142 PD-L1 positive. SP142 PD-L1 positivity was significantly associated with tumor-infiltrating lymphocytes (P <0.01), and node negativity (P=0.02), but not with tumor grade (P=0.35), tumor size (P=0.58), or BRCA mutation (P=0.53). Overall percentage agreement (OPA) for intraobserver and interobserver agreement was 95.0% and 93.7%, respectively, among 5 pathologists trained in TNBC SP142 PD-L1 scoring. In 5 TNBC SP142 PD-L1-naive pathologists, significantly higher OPA to the reference score was achieved after video training (posttraining OPA 85.7%, pretraining OPA 81.5%, P<0.05). PD-L1 status at a 1% cutoff was assessed by SP142 and SP263 in 420 cases, and by SP142 and 22C3 in 423 cases, with OPA of 88.1% and 85.8%, respectively. The VENTANA PD-L1 (SP142) Assay is reproducible for classifying TNBC PD-L1 status by trained observers; however, it is not analytically equivalent to the VENTANA PD-L1 (SP263) Assay and Dako PD-L1 IHC 22C3 pharmDx assay.
Triple-negative breast carcinoma (TNBC) is an aggressive subtype of breast carcinoma, traditionally with limited treatment options, but is one of an increasing number of tumor types that may respond to immunotherapy. As with other tumor types treated with immunotherapy, particularly programmed cell death protein 1 (PD-1) and programmed cell death ligand 1 (PD-L1) inhibitors, eligibility is based on companion diagnostic immunohistochemical assays. These PD-L1 assays are specific to tumor type and therapeutic agent, require specific laboratory platforms, and have specific scoring systems and positivity thresholds for tumor and immune cells or a combination thereof.1
The recently published clinical trial IMpassion130 (NCT02425891) showed significantly longer progression-free survival and overall survival with the anti-PD-L1 antibody atezolizumab in combination with nab-paclitaxel compared with placebo and nab-paclitaxel in SP142 PD-L1-positive (≥1% immune cell positivity) metastatic and locally advanced, unresectable TNBC.2 The VENTANA PD-L1 (SP142) Assay (abbreviated to SP142) is the designated companion diagnostic test for atezolizumab, therefore accurate assessment of this assay is essential for identifying patients likely to benefit from atezolizumab.
However, there are limited data on interpathologist and intrapathologist agreement for SP142 PD-L1 assessment in TNBC. One study reported high interpathologist and intrapathologist agreement in the classification of SP142 PD-L1 status at a 1% cutoff in a limited number of breast carcinoma cases (n=30) and pathologists (n=3),3 whereas another reported an interclass correlation coefficient of 0.560 for classification of SP142 PD-L1 status at a 1% cutoff in a cohort of 68 TNBCs by 19 pathologists without prior specific training in this area.4 It is not stated whether cases in these 2 studies were enriched around the clinically critical cutpoint of 1%, although the latter study reports a positivity rate of 58% with a mean score of 20% in the positive cases, suggesting a skew towards higher scores.4 The only published study investigating SP142 PD-L1 interobserver agreement in TNBCs enriched around the 1% cutpoint reported an interclass correlation coefficient of 0.805 among 7 specifically trained pathologists in a limited cohort of cases (n=30).5
Therefore the true reproducibility by pathologists trained in SP142 PD-L1 assessment in TNBC, and the effect of training in determining PD-L1 positivity in cases close to the critical decision point of 1%, remains largely unknown.
Furthermore, there are no substantial data regarding concordance between the absolute percentages or clinical cutpoints in comparing the different anti-PD-L1 antibody clones and their specific instrumentation.
The existing studies addressing this issue in TNBC have been on small case cohorts including up to 196 cases only.3–7 There is only one study on a larger population of the magnitude of our study (n=420) which was the IMpassion130 study population.8
Data on interassay concordance is essential because in-house PD-L1 testing is likely to be impracticable for the majority of anatomic pathology laboratories that utilize a single immunohistochemical platform if the clones and platforms are not interchangeable. Thus, addressing whether PD-L1 assays currently used for other common tumor types but not Food and Drug Administration (FDA)-approved for TNBC, such as the VENTANA PD-L1 (SP263) Assay (abbreviated to SP263) for urothelial carcinoma, and Dako PD-L1 IHC 22C3 pharmDx assay (abbreviated to 22C3) for non–small cell lung carcinoma, gastroesophageal junction, urothelial and cervical carcinomas, could be substituted for the SP142 assay to determine PD-L1 status in TNBC is critically important.
Therefore, the aims of this study were: (1) assess the prevalence of SP142 PD-L1 positivity and association of SP142 PD-L1 positivity with clinicopathologic features in the Australian TNBC population, (2) to determine the intraobserver and interobserver agreement in pathologists trained in TNBC SP142 PD-L1 assessment, (3) determine the effect of training on accuracy of SP142 PD-L1 scoring by pathologists naïve for SP142 PD-L1 scoring, and (4) determine the concordance between the SP142, SP263, and 22C3 PD-L1 assays in TNBC.
MATERIALS AND METHODS
Tissue Samples
Fifteen tissue microarrays (TMAs) were constructed using a total of 1133 cores, 1 mm in diameter, from 562 previously untreated, resected primary invasive breast carcinomas with a triple-negative (estrogen receptor–negative, progesterone receptor–negative, human epidermal growth factor receptor 2–negative) phenotype. The cases were derived from 3 hospital sites, 1 private pathology laboratory, and 1 familial cancer consortium to capture a range of TNBCs—Peter MacCallum Cancer Centre (PMCC), TissuPath (TP) (combined PMCC and TP, n=286, tumors resected 2000-2018), Concord Repatriation General Hospital (CRGH, n=104, tumors resected, 1997-2013), Royal Prince Alfred Hospital (RPAH, n=88, tumors resected, 1995-2010), and Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) (n=84, patients with known BRCA mutation status, tumors resected, 1980-2008). To achieve an adequately sized cohort, it was necessary to include samples older than originally intended (<10 y). A total of 199 tumors were represented by >1 core (2 to 6 cores) in the TMAs.
The clinicopathologic details of the cases were obtained from surgical pathology reports, and the triple-negative phenotype of the tumors was confirmed in the CRGH, RPAH, and kConFab cohorts by repeat immunohistochemistry for estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 before TMA construction. Tumor-infiltrating lymphocytes (TILs) were scored on whole sections and core biopsies of the tumor according to guidelines published by the International TILs Working Group,9 expressed as a percentage of tumor-associated stroma occupied by TILs and further categorized as 0 (virtually absent), 1 (mild, <30%), 2 (moderate, 30% to 60%), and 3 (marked, >60%).10,11
This project was approved by the human research ethics committee of Peter MacCallum Cancer Centre (project 03/90).
PD-L1 Immunohistochemistry
Serial sections of the TMAs were cut at 4 µm thickness and immunohistochemistry for PD-L1 clones SP142, SP263, and 22C3 was performed at PMCC within 3 weeks of sectioning (PMCC/TP and kConFab TMAs) or within 2 months of sectioning (RPAH and CRGH TMAs). Immunohistochemistry for SP142, SP263, and 22C3 PD-L1 was performed using locked protocols for the CE-IVD PD-L1 kits on the Ventana BenchMark ULTRA Platform (SP142 and SP263) and the Dako Link 48 platform (22C3). The instrument performed the staining process by applying the appropriate reagent, monitoring the incubation time and rinsing slides between reagents. Omission of the primary antibody was used as a negative control. Tissue samples were subsequently counterstained with hematoxylin and mounted in nonaqueous, permanent mounting media. Appropriately stained external controls comprising tonsil and placenta were present on each TMA section.
Scoring of PD-L1 Immunohistochemistry
Full-face cores containing at least 100 invasive carcinoma cells (determined by manual counting) were required for assessment for PD-L1 status. For each PD-L1 clone, up to 15.7% and 13.2% of cores were discarded due to insufficient tumor cells or partial sections, respectively. PD-L1 scores were expressed as the percentage of tumor area occupied by positive-stained immune cells.12 The PD-L1 scores were categorized as tumor infiltrating immune cells (IC) 0 (<1%), IC 1(1% to <5%), IC 2 (5% to <10%), and IC 3 (at least 10%) (Fig. 1), and dichotomized as PD-L1 negative (<1%) or PD-L1 positive (≥1%). Scoring was performed by 2 pathologists (J.-M.B.P. and S.B.F.) who were trained and demonstrated competency in SP142 PD-L1 assessment in TNBC in a 1-day training course, and were experienced in SP142, SP263, and 22C3 PD-L1 assessment in clinical samples. All the cores were scored for SP142, SP263, and 22C3 PD-L1 by 1 investigator (J.-M.B.P.). Where there were multiple cores from the same tumor, the highest PD-L1 score was taken. Cores scored adjacent to the cutpoint (<1% to 5%) for SP142 PD-L1, and cores with discordant PD-L1 status between the PD-L1 assays were double scored with another pathologist (S.B.F.). Overall, 62.9% (281/447) of SP142 PD-L1 scores, 35.9% (166/462) of SP263 PD-L1 scores, and 35.2% (159/452) of 22C3 PD-L1 scores were reviewed. All 60 cores included in the intraobserver and interobserver reproducibility study were scored by both pathologists to generate the reference score.
FIGURE 1.
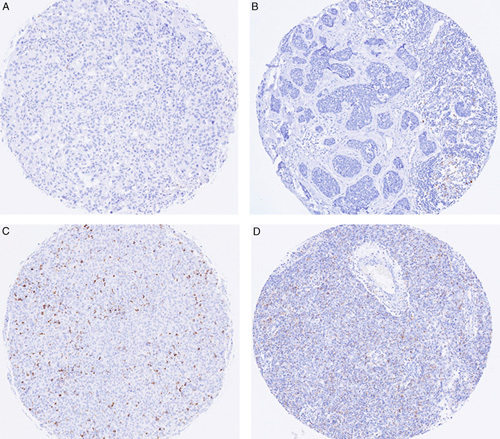
SP142 PD-L1 staining of inflammatory cells in tumor-associated stroma. A, <1%. B, 1% to <5%. C, 5% to <10%. D, ≥10%.
Intraobserver and Interobserver Reproducibility and Impact of Training on SP142 PD-L1 Assessment
The number of pathologists included in this part of the study was determined based on a statistical power calculation from an expected true overall percent agreement (OPA) of 89% for intraobserver and interobserver concordance, and it was calculated that 5 pathologists were required for each subgroup to generate 300 pairwise comparisons to ensure the lower bound of the Wilson 95% confidence interval (CI) of OPA to be >85%. The 2 subgroups of 5 pathologists each scored a cohort of 60 cases on 2 consecutive days with an overnight washout period, to generate 300 pairwise comparisons for each subgroup on each day (Fig. 2). To evaluate the reproducibility of SP142 PD-L1 status assessment in a clinically relevant manner, the 60 selected cases of the cohort were enriched around the 1% cutoff, with 29 negative samples (0% or <1%), 21 positive samples close to the 1% threshold (1% to 10%), and 10 positive samples far from the 1% threshold (>10%). The 60 TMA cores were distributed over 12 slides with 1 to 6 cores for assessment on each slide and each slide was scored in a random order. On each day, the participating pathologists assessed the same 60 SP142 PD-L1-stained tumor cores for PD-L1 status, recording scores for each case on a pro forma study response form (Supplementary Material, Supplemental Digital Content 1, http://links.lww.com/PAS/B95).
FIGURE 2.
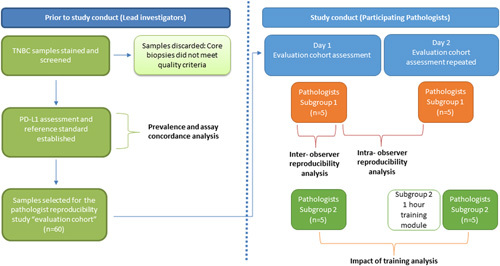
Outline of study.
The 10 pathologists represented a range of clinical practice and experience throughout Australia. Five of the pathologists were trained and had demonstrated competency during a 1-day training course in SP142 PD-L1 assessment of TNBC 7 months before the study (subgroup 1) and 5 pathologists were selected as they were untrained in SP142 PD-L1 assessment in TNBC (subgroup 2). The subgroup 2 pathologists were only given the scoring criteria before assessment on the first day and watched a 1-hour long instructional video and received an interpretation guide on TNBC SP142 PD-L1 assessment before their assessment of the cases on the second day.
Intraobserver reproducibility was evaluated by comparing the scores obtained by subgroup 1 on days 1 and 2. Interobserver reproducibility was evaluated by comparing the scores between subgroup 1 pathologists on day 1.
The impact of training was assessed by comparing the scores obtained by the 5 previously untrained pathologists (subgroup 2) before (day 1) and after (day 2) receiving SP142 PD-L1 assessment in TNBC training.
In addition, exploratory analyses of interobserver reproducibility between 4, 6, 8, and 10 pathologists, each composed of equal numbers from subgroups 1 and 2, were also performed to assess the impact of observer numbers on interobserver agreement.
Statistical Design and Analysis
To estimate the prevalence of SP142 PD-L1-positive TNBCs to a precision of 5% (ie, the exact Clopper-Pearson 95% CI was no >±5%), it was determined that 369 TNBC cases were required, based on a previously reported positivity rate of ∼40% in the TNBC population.2 For the interobserver and intraobserver concordance study, with an expected OPA of 89% for both interobserver and intraobserver agreement, it was estimated that a total of 300 pairwise comparisons were required for both the interobserver and intraobserver assessments of agreement, for the lower limit of the Wilson 95% CI of the OPA to be >85%. To assess analytical concordance between PD-L1 assays, for an OPA between 2 assays of 80% and ensuring the lower limit of the 95% CI was at least 75% using Wilson CIs, it was determined that at least 290 samples were required. Interobserver and intraobserver reproducibility was assessed using OPA, average positive agreement (APA), and average negative agreement (ANA). Cohen κ coefficient and prevalence-adjusted bias-adjusted kappa (PABAK) were calculated. Analytical concordance between the different PD-L1 assays was assessed using OPA, positive percent agreement (PPA), and negative percent agreement (NPA). 95% CIs were computed for all measurements. Association of SP142 PD-L1 status with categorical variables was assessed using the Fisher exact test and χ2 test. Statistical analyses were undertaken using SAS software, version 9.4.
RESULTS
Cohort Characteristics
The TMAs contained cores from 562 triple-negative invasive breast carcinoma cases. All the patients were female. The clinicopathologic characteristics of the cohort are summarized in Table 1. TILs scores were available on 293 tumors, and as a continuous variable in 134 cases. Where TILs were expressed as a continuous variable, TILs ranged from 0% to 100% (median: 15%). TILs were categorized as score 0 in 20.3% of tumors, score 1 in 48.1%, score 2 in 21.7%, and score 3 in 10.0% of tumors. There was a significant difference in TILs scores between the 4 source sites, with TILs classified as virtually absent in 25.6% and 38.4% of tumors from CRGH and RPAH, respectively, compared with only 5.7% and 8.1% of cases from PMCC/TP and kConFab, respectively (P<0.0001).
TABLE 1.
Cohort Characteristics
| n (%) | |
|---|---|
| Age (n=366), median (range) (y) | 59 (23-96) |
| BRCA status (n=84) | |
| BRCA1 | 42 (50.0) |
| BRCA2 | 12 (14.3) |
| BRCAX | 30 (35.7) |
| Tumor size (n=382) (mm) | |
| ≤20 | 165 (43.2) |
| >20 to ≤50 | 191 (50.0) |
| >50 | 26 (6.8) |
| Median (range) (mm) | 22 (0.7-220) |
| Tumor grade (n=342) | |
| Grade 1 | 5 (1.5) |
| Grade 2 | 32 (9.4) |
| Grade 3 | 305 (89.2) |
| Tumor type (n=243) | |
| Infiltrating ductal carcinoma | 217 (89.3) |
| Invasive carcinoma with medullary phenotype | 11 (4.5) |
| Invasive lobular carcinoma | 5 (2.1) |
| Other | 10 (4.1) |
| Carcinoma NOS; adenocarcinoma NOS (n) | 2 |
| Infiltrating ductal and lobular carcinoma (n) | 2 |
| Metaplastic carcinoma (n) | 1 |
| Papillary carcinoma (n) | 1 |
| Secretory carcinoma (n) | 1 |
| Tubular adenocarcinoma (n) | 1 |
| Apocrine carcinoma (n) | 1 |
| Poorly differentiated carcinoma with neuroendocrine features (n) | 1 |
| Nodal status (n=335) | |
| pN0 | 201 (60.0) |
| pN1 | 90 (26.9) |
| pN2 | 23 (6.9) |
| pN3 | 21 (6.3) |
NOS indicates not otherwise specified.
There was no statistically significant correlation between TIL scores and tumor grade, tumor size, lymph node status, and disease stage (P>0.05, data not shown).
Prevalence of SP142 PD-L1 Positivity
Cores from 447 tumors were of sufficient quality to be scored for SP142 PD-L1. Scores ranged from 0% to 60% (median score: <1%). At a 1% cutoff, 34.2% were positive for the SP142 assay, with 60.8% of the positive cases scoring <5%. The prevalence of SP142 PD-L1 positivity, as scored by the 2 principal pathologists (J.-M.B.P., S.B.F.), varied between the source sites, 42.9%, 36.1%, 32.4%, and 25.8% of cases were positive from kConFab, PMCC/TP, RPAH, and CRGH, respectively (Table 2). In the 46 cases where >1 core from a single tumor was assessed for SP142 PD-L1 status, there was discordant classification of the tumor as PD-L1 positive or negative in 25 (54.4%) cases, indicating heterogeneity of SP142 PD-L1 staining within individual tumors. There was no statistically significant difference in tumor grade, tumor size, lymph node status, and disease stage between tumors demonstrating PD-L1 heterogeneity and tumors without PD-L1 heterogeneity (P>0.05, data not shown).
TABLE 2.
Prevalence of PD-L1 Positivity (at a 1% Cutoff) by Source Site and PD-L1 Clone
| Prevalence Of PD-L1 Positivity, n/N (%) | |||||
|---|---|---|---|---|---|
| Total, n/N (%) | PMCC/TP | kConFab | RPAH | CRGH | |
| SP142 | 153/447 (34.2) | 86/238 (36.1) | 21/49 (42.9) | 23/71 (32.4) | 23/89 (25.8) |
| SP263 | 197/462 (42.5) | 106/241 (4.0) | 26/65 (40.0) | 31/65 (47.7) | 34/91 (37.4) |
| 22C3 | 159/452 (35.2) | 110/237 (46.4) | 19/50 (38.0) | 15/70 (21.4) | 15/95 (15.8) |
SP142 PD-L1 Status Is Associated With TILs and Nodal Status
There was an increased frequency of SP142 PD-L1 positivity with increasing density of TILs (P<0.001, Table 3). Node negative primary breast tumors were more frequently SP142 PD-L1 positive compared with primary tumors with lymph node metastases (P=0.02, Table 3). There was no significant association between SP142 PD-L1 status and tumor grade (P=0.35), tumor size (P=0.58), or BRCA mutation status (P=0.53) (Table 3).
TABLE 3.
SP142 PD-L1 Status and Tumor Features
| SP142 PD-L1 Status | |||
|---|---|---|---|
| SP142 PD-L1 Negative | SP142 PD-L1 Positive | P | |
| Tumor grade | |||
| Grade 1 | 2 | 2 | 0.35 |
| Grade 2 | 15 | 4 | |
| Grade 3 | 153 | 85 | |
| Tumor size (mm) | |||
| ≤20 | 79 | 45 | 0.58 |
| >20 to ≤50 | 105 | 50 | |
| >50 | 11 | 8 | |
| Nodal status | |||
| pN0 | 97 | 63 | 0.017 |
| pN1-N3 | 79 | 26 | |
| TILs | |||
| Virtually absent | 43 | 6 | <0.0001 |
| Mild, <30% | 81 | 28 | |
| Moderate, 30% to ≤60% | 22 | 26 | |
| Marked, >60% | 5 | 13 | |
| BRCA status | |||
| BRCA1 | 12 | 12 | 0.53 |
| BRCA2 | 4 | 3 | |
| BRCAX | 12 | 6 | |
Intraobserver and Interobserver Reproducibility Among Pathologists With Specific Training in SP142 PD-L1 Assessment in TNBC Is High
For subgroup 1 pathologists, pairwise comparisons of day 1 and day 2 results for each of the 60 evaluated samples showed excellent intraobserver agreement, with an OPA of 95.0% (95% CI: 91.9%-97.0%), APA of 95.2% (95% CI: 92.2%-97.0%), and ANA of 94.9% (95% CI: 91.7%-96.9%). Cohen κ coefficient was 0.9 (almost perfect strength of agreement, 95% CI: 0.9-1.0) and PABAK was 0.9.
Interobserver agreement between subgroup 1 pathologists (day 1) was also excellent, with an OPA of 93.3% (95% CI: 91.1%-95.2%), APA of 93.6% (95% CI: 91.5%-95.3%), and ANA of 93.0% (95% CI: 90.6%-94.8%). Cohen κ coefficient was 0.9 (almost perfect strength of agreement, 95% CI: 0.8-0.9) and PABAK was 0.9.
Training in TNBC SP142 PD-L1 Assessment Improves Accuracy of Assessment
SP142 PD-L1 scores by pathologists in subgroup 2 were compared with the reference score on day 1 (before SP142 PD-L1 assessment in TNBC training) and on day 2 (after training). Following training, there was an improvement in OPA (day 1 OPA: 81.5%, 95% CI: 76.8%-85.5%; day 2 OPA: 85.7%, 95% CI: 81.3%-89.2%, P<0.05). However, OPA of day 2 scores to the reference score was still higher in subgroup 1 pathologists who had previously participated in the longer training course (OPA: 96.3%, 95% CI: 93.6%-97.9%) compared with subgroup 2 pathologists who received 1 hour of training on day 2 (OPA 85.7%, 95% CI: 81.3%-89.2%).
Subgroup 2 interpathologist agreement also improved from an OPA of 76.0% (95% CI: 72.4%-79.3%), Cohen κ coefficient 0.5 (moderate strength of agreement, 95% CI: 0.4-0.6) on day 1 to an OPA of 81.3% (95% CI: 78.0%-84.3%), Cohen κ coefficient 0.6 (substantial strength of agreement, 95% CI: 0.5-0.7) following training on day 2.
Exploratory analyses of interobserver reproducibility between 4, 6, 8, and 10 pathologists, each composed of equal numbers from subgroups 1 and 2, were also performed to assess the impact of observer numbers on interobserver agreement.
Interobserver agreement was consistent on each day of the study for all group sizes (day 1 OPA: 81.6%, 82.4%, 83.0%, 83.2% for 4, 6, 8, and 10 pathologists, respectively; day 2 OPA: 88.3%, 85.8%, 84.8%, 85.9% for 4, 6, 8, and 10 pathologists, respectively).
SP142, SP263, and 22C3 PD-L1 Assays Are Not Analytically Equivalent in TNBC
A total of 462 and 452 cases were of sufficient quality to be scored for SP263 PD-L1, and 22C3 PD-L1, respectively. PD-L1 status was available for all 3 clones in 403 cases, SP142 and SP263 in 420 cases, SP142 and 22C3 in 423 cases, and for SP263 and 22C3 in 422 cases.
At a 1% cutoff, 42.6% were positive for the SP263 assay, and 35.2 were positive for the 22C3 assay. The prevalence of 22C3 PD-L1 positivity varied between the source sites, ranging between 15.8% (CRGH) and 46.4% (PMCC/TP), while the prevalence of SP263 PD-L1 positivity was more constant, ranging between 37.4% and 44.0% between the cohorts (Table 2).
Comparing PD-L1 status assessed by SP142 and SP263 at a 1% cutoff (Fig. 3), OPA was 88.1% (95% CI: 85.0%-91.2%), PPA 95.2% (95% CI: 91.7%-98.7%), and NPA 84.3% (95% CI: 80.0%-88.6%). For SP142 and 22C3 (Fig. 3), OPA was 85.8% (95% CI: 82.5%-89.1%), PPA 81.4% (95% CI: 75.0%-87.7%), and NPA 88.1% (95% CI: 84.3%-91.9%).
FIGURE 3.

Concordance between PD-L1 status assessed by SP142, SP263, and 22C3 clones (Venn diagrams are not to scale). A, SP142 and 22C3 PD-L1 positivity at a 1% cutoff in 423 cases where both SP142 and 22C3 PD-L1 status was assessable. B, SP142 and SP263 PD-L1 positivity at a 1% cutoff in 420 cases where both SP142 and 22C3 PD-L1 status was assessable. C, SP142, 22C3, and SP263 PD-L1 positivity at a 1% cutoff in 403 cases where PD-L1 status by all 3 clones was assessable.
DISCUSSION
Several studies have demonstrated significantly improved outcomes with the addition of anti-PD-L1 therapy to chemotherapy in patients with TNBC showing at least 1% SP142 PD-L1 immune cell expression2,13,14 with durable effect,2,14,15 emphasizing the importance of accurate assessment of PD-L1 status in TNBCs. This is one of the first studies to investigate the reproducibility of PD-L1 assessment using the VENTANA PD-L1 (SP142) Assay in TNBCs and was specifically designed to have sufficient statistical power for meaningful analysis, and with samples enriched around the 1% cutoff for designation of SP142 PD-L1 positivity. Similar to other studies,13,16,17 SP142 PD-L1 immune cell positivity was observed in all tumor stages, and unsurprisingly was significantly associated with high stromal TILs. SP142 PD-L1 immune cell positivity in the primary tumor was significantly associated with the absence of nodal disease. In TNBCs, tumor cell PD-L1 expression has been reported to be inversely associated with lymph node involvement using the SP14218 and 28-819 PD-L1 clones, and stromal SP142 PD-L1 expression at a 1% cutoff to be associated with the absence of lymphovascular space invasion.20 Higher prevalence of SP142 PD-L1 positivity in high-grade tumors was not observed in our cohort, possibly due to the relatively small number of non–grade 3 tumors in this study. No significant association of SP142 PD-L1 expression with BRCA1/2 mutation status was observed, similar to other published studies using the SP14216 and other PD-L1 antibodies.21,22
Overall, 34% of TNBCs in this study were PD-L1 positive as assessed by the SP142 PD-L1 assay at a 1% cutoff, less than the prevalence of 41% reported in the IMpassion130 trial.2 We did observe differences in prevalence of positivity for different clones in the different source cohorts, suggesting that significant differences in TILs between tumors from the source sites, as well as other possible unquantified differences in populations, age of the tumor tissue or time from sectioning to staining (the latter recognized to lead to the loss of staining in older samples12,23), may have influenced the observed prevalence of PD-L1 positivity.
The use of TMAs in our study might have contributed to an underestimation of PD-L1-positive cases, especially where there is heterogeneity within an individual tumor. This is seen in this study at ∼50% and by others at up to 50%16 although this might be more prevalent in metastatic sites where the overall frequency of PD-L1 positivity has been reported to be lower.8 Nevertheless, the use of TMAs in this study allowed standardization of staining across many samples and ensured the same area of tumor was assessed for intraobserver and interobserver reproducibility and for comparison of the PD-L1 assays. TMAs have been demonstrated to be appropriate for investigation of both PD-L1 assay concordance3,6,19,24–27 and PD-L1 intraobserver and interobserver concordance3,26,28 in breast carcinoma and other tumor types. In addition, compared with whole tissue sections, TMA cores more closely mimic the metastatic disease setting where small biopsies are the norm and PD-L1 assessment is most clinically relevant.
There was an almost perfect intraobserver and interobserver agreement and agreement to the reference score in assessment of PD-L1 status using the SP142 assay among pathologists who received specific training in SP142 PD-L1 assessment in TNBC through a day-long training seminar.
Among untrained pathologists, we observed higher agreement to the reference score and interobserver agreement than that previously reported,4 but below the level of agreement of 85% suggested to be acceptable for semiquantitative assays.29 An hour-long training video improved SP142 PD-L1 agreement to the reference score and interpathologist agreement.
The importance of specific training in SP142 PD-L1 assessment is supported by a recent study by Reisenbichler et al4 which found only 38% concordance for SP142 PD-L1 status in TNBCs at a 1% cutoff among pathologists who had received minimal training before scoring, although the increased complexity of assessing SP142 PD-L1 status on whole sections, given the known heterogeneity of SP142 PD-L1 staining in TNBCs, and the use of scanned slides rather than glass slides may at least partly account for the lower interpathologist agreement. This same study also showed that the number of observers required for the interobserver agreement to plateau is inversely proportional to the robustness of the assay, and for the TNBC SP142 PD-L1 assay, interobserver agreement plateaus at 9 pathologists. In contrast, we observed consistent levels of interobserver agreement on each day of the study between 4, 6, 8, and 10 pathologists composed of equal numbers from each subgroup. This suggests that formal training and experience in TNBC SP142 PD-L1 assessment, like many other aspects of diagnostic surgical pathology,30 is an important determinant of interpathologist agreement rather than pathologist number alone.
The study was also designed to assess whether the different assays were interchangeable. Although SP142 is the only assay designed to identify immune cell reactivity alone, and the SP263 and 22C3 assays are not typically scored as a percentage of tumor area occupied by positive-staining inflammatory cells in their role as companion diagnostic assays in other tumor types, this scoring method was used in this study to directly compare the 3 assays. At a 1% cutoff, we observed different prevalences of PD-L1 positivity, the highest being the SP263 assay, followed by the 22C3 assay. The SP142 assay had the lowest prevalence of PD-L1 positivity. This pattern is consistent with our and others’ observations in lung cancer25,31 and with a recent meta-analysis of PD-L1 assay concordance in a range of tumor types, including non–small cell lung carcinoma, urothelial carcinoma, mesothelioma, and thymic carcinoma. In the latter study, the SP263 assay was the most sensitive of Food and Drug Administration (FDA)-approved PD-L1 assays, and the SP142 assay showed lower sensitivity compared with the 22C3 and the SP263 assays.32
In our study population, PD-L1 assay substitution would result in 6.4% and 1.7% of SP142 PD-L1-positive patients being classified as PD-L1 negative by the 22C3 assay and SP263 assay, respectively. Conversely, 7.8% and 10.2% of SP142 PD-L1 negative patients would be classified as PD-L1 positive by the 22C3 and SP263 assays, respectively.
Thus, as reported in other tumor types,25,32–34 these findings indicate that the commercially available SP142, SP263, and 22C3 assays are not analytically equivalent in TNBCs as defined by an OPA of at least 90%.35 Our findings are consistent with those from the IMpassion130 study which reported OPAs of 69% for SP142 and 22C3, and 63% for SP142 and SP263,8 and with the lack of analytical equivalence between SP142 and SP263 PD-L1 clones observed by Reisenbichler et al4 and by Scott et al,7 and with the recent study by Noske et al5 which reported 10% disagreement in PD-L1 status between SP142 and 22C3 and over 20% disagreement between SP142 and SP263. This may in part due to the different epitopes targeted by the clones as 22C3 targets the extracellular domain of PD-L1, while SP142 targets an epitope on the cytoplasmic domain of PD-L1. However, SP142 and SP263 target an identical epitope, suggesting that this is not the only cause for the observed differences in PD-L1 staining.36
Whether classification of PD-L1 status by the different assays results in equivalent clinical outcomes is unknown as there is a paucity of clinical trials to specifically address this question. Nevertheless, there is evidence from IMpassion130 that using the different clones 22C3 and SP263 stratified by combined positive score of 1 and IC staining at the 1% cutpoint respectively, that patients show similar differences in outcome when treated with atezolizumab, although they are not precisely the same patients.8
The need for a specific PD-L1 clone, platform, and interpretation method for different tumor types presents a practical problem for many pathology laboratories but might be necessary for appropriate treatment decisions given the significant costs and potential adverse effects of immunotherapy agents. It also perhaps argues for this type of biomarker testing to be performed centrally rather than in small laboratories that might report few cases, as it is recognized that volume of reporting for many aspects of pathology significantly influences accuracy and reproducibility.37–39
In summary, this study demonstrates the VENTANA PD-L1 (SP142) Assay to have excellent intraobserver and interobserver reproducibility among pathologists with specific, detailed training in SP142 PD-L1 assessment in TNBC, but lower agreement among pathologists untrained or with minimal training in SP142 PD-L1 assessment in TNBC. The VENTANA PD-L1 (SP142) Assay, VENTANA PD-L1 (SP263) Assay, and Dako PD-L1 IHC 22C3 pharmDx assay are not analytically equivalent in the assessment of PD-L1 status in TNBC.
Supplementary Material
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.ajsp.com.
ACKNOWLEDGMENTS
The authors thank Heather Thorne, Eveline Niedermayr, Sharon Guo, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health [US]) for their contributions to this resource, and the many families who contribute to kConFab.
Footnotes
Conflicts of Interest and Source of Funding: Supported by Roche Products Pty. Limited (Australia) who is the study sponsor. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia, and the Cancer Foundation of Western Australia. S.B.F. is funded by the NHMRC Practitioner Fellowship (APP 1079329). S.A.O.T. is funded by the National Breast Cancer Foundation (PRAC 16-006 and IIRS-19-84) and the Sydney Breast Cancer Foundation. S.A.O.T. has received travel/accommodation support from Roche for study participation and has received honorarium for participation in Advisory Boards from Roche, BMS and Merck. B.C. is a full-time employee of Roche. P.B. is an employee of OzBiostats Pty. Limited, who were contracted by Roche to work on the study. J.A., W.A.C., E.K.A.M., W.R., and V.S. have received travel/accommodation support from Roche for study participation. S.R.L. has received honorarium for participation in the Roche Advisory Board (2020) on HER2 and TNBC; is a member of the board of directors Breast Cancer Trials (formerly ANZ Breast Cancer Trial Group); and has received travel/accommodation support from Roche for study participation. J.B. has received honorarium and travel/accommodation support from Roche. For the remaining authors none were declared.
Contributor Information
Jia-Min B. Pang, Email: jia-min.pang@petermac.org.
Belinda Castles, Email: belinda.castles@roche.com.
David J. Byrne, Email: david.byrne@petermac.org.
Peter Button, Email: peter@ozbiostat.com.
Shona Hendry, Email: shona.hendry@gmail.com.
Sunil R. Lakhani, Email: s.lakhani@uq.edu.au.
Vanathi Sivasubramaniam, Email: vanathi.sivasubramaniam@svha.org.au.
Wendy A. Cooper, Email: Wendy.Cooper@health.nsw.gov.au.
Jane Armes, Email: Jane.Armes@health.qld.gov.au.
Ewan K.A. Millar, Email: Ewan.Millar@SESIAHS.HEALTH.NSW.GOV.AU.
Wendy Raymond, Email: WRaymond@clinpath.com.au.
Samuel Roberts-Thomson, Email: Samuel.RobertsThomson@mh.org.au.
Beena Kumar, Email: Beena.Kumar@monashhealth.org.
Marian Burr, Email: Marian.Burr@mh.org.au.
Christina Selinger, Email: tinas@rcpa.edu.au.
Kate Harvey, Email: k.harvey@garvan.org.au.
Charles Chan, Email: Charles.Chan2@health.nsw.gov.au.
Jane Beith, Email: jane.beith@lh.org.au.
David Clouston, Email: david.clouston@tissupath.com.au.
Sandra A. O’Toole, Email: Sandra.OToole@health.nsw.gov.au.
Stephen B. Fox, Email: stephen.fox@petermac.org.
Collaborators: kConFab
REFERENCES
- 1.Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer. 2019;7:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379:2108–2121. [DOI] [PubMed] [Google Scholar]
- 3.Downes MR, Slodkowska E, Katabi N, et al. Inter- and intraobserver agreement of programmed death ligand 1 scoring in head and neck squamous cell carcinoma, urothelial carcinoma and breast carcinoma. Histopathology. 2020;76:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reisenbichler ES, Han G, Bellizzi A, et al. Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer. Mod Pathol. 2020;33:1746–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noske A, Ammann JU, Wagner DC, et al. A multicentre analytical comparison study of inter-reader and inter-assay agreement of four programmed death-ligand 1 (PD-L1) immunohistochemistry assays for scoring in triple-negative breast cancer. Histopathology. 2021;78:567–577. [DOI] [PubMed] [Google Scholar]
- 6.Lee SE, Park HY, Lim SD, et al. Concordance of programmed death-ligand 1 expression between SP142 and 22C3/SP263 assays in triple-negative breast cancer. J Breast Cancer. 2020;23:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott M, Scorer P, Barker C, et al. Comparison of patient populations identified by different PD-L1 assays in triple-negative breast cancer (TNBC). Ann Oncol. 2019;(30 suppl 3):iii1– iii26. [Google Scholar]
- 8.Rugo HS Loi S Adams S, et al. Performance of PD-L1 immunohistochemistry (IHC) assays in unresectable locally advanced or metastatic triple-negative breast cancer (MTNBC): post-hoc analysis of IMpassion130. ESMO 2019 Congress. Barcelona, Spain: Annals of Oncology; 2019:v851–v934.
- 9.Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schalper KA, Velcheti V, Carvajal D, et al. In situ tumor PD-L1 mRNA expression is associated with increased TILs and better outcome in breast carcinomas. Clin Cancer Res. 2014;20:2773–2782. [DOI] [PubMed] [Google Scholar]
- 11.Beckers RK, Selinger CI, Vilain R, et al. Programmed death ligand 1 expression in triple-negative breast cancer is associated with tumour-infiltrating lymphocytes and improved outcome. Histopathology. 2016;69:25–34. [DOI] [PubMed] [Google Scholar]
- 12.Diagnostics R. Ventana PD-L1 (SP142) Assay Interpretation Guide for Triple-Negative Breast Carcinoma (TNBC). North Ryde, NSW, Australia: Roche Diagnostics Australia Pty. Limited; 2019. [Google Scholar]
- 13.Cerbelli B, Pernazza A, Botticelli A, et al. PD-L1 expression in TNBC: a predictive biomarker of response to neoadjuvant chemotherapy? Biomed Res Int. 2017;2017:1750925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emens LA, Cruz C, Eder JP, et al. Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: a phase 1 study. JAMA Oncol. 2019;5:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid P Adams S Rugo HS, et al. IMpassion130: updated overall survival (OS) from a global, randomized, double-blind, placebo-controlled, phase III study of atezolizumab (atezo)+nab-paclitaxel (nP) in previously untreated locally advanced or metastatic triple-negative breast cancer (mTNBC). Chicago, IL; ASCO Annual Meeting 2019; 2019.
- 16.Dill EA, Gru AA, Atkins KA, et al. PD-L1 expression and intratumoral heterogeneity across breast cancer subtypes and stages: an assessment of 245 primary and 40 metastatic tumors. Am J Surg Pathol. 2017;41:334–342. [DOI] [PubMed] [Google Scholar]
- 17.Stovgaard ES, Bokharaey M, List-Jensen K, et al. PD-L1 diagnostics in the neoadjuvant setting: implications of intratumoral heterogeneity of PD-L1 expression in triple negative breast cancer for assessment in small biopsies. Breast Cancer Res Treat. 2020;181:553–560. [DOI] [PubMed] [Google Scholar]
- 18.Botti G, Collina F, Scognamiglio G, et al. Programmed death ligand 1 (PD-L1) tumor expression is associated with a better prognosis and diabetic disease in triple negative breast cancer patients. Int J Mol Sci. 2017;18:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun WY, Lee YK, Koo JS. Expression of PD-L1 in triple-negative breast cancer based on different immunohistochemical antibodies. J Transl Med. 2016;14:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HS, Do SI, Kim DH, et al. Clinicopathological and prognostic significance of programmed death ligand 1 expression in Korean patients with triple-negative breast carcinoma. Anticancer Res. 2020;40:1487–1494. [DOI] [PubMed] [Google Scholar]
- 21.Sobral-Leite M, Van de Vijver K, Michaut M, et al. Assessment of PD-L1 expression across breast cancer molecular subtypes, in relation to mutation rate, BRCA1-like status, tumor-infiltrating immune cells and survival. Oncoimmunology. 2018;7:e1509820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solinas C, Marcoux D, Garaud S, et al. BRCA gene mutations do not shape the extent and organization of tumor infiltrating lymphocytes in triple negative breast cancer. Cancer Lett. 2019;450:88–97. [DOI] [PubMed] [Google Scholar]
- 23.Tsao MS, Kerr KM, Dacic S, et al. IASLC Atlas of PD-L1 Immunohistochemistry Testing in Lung Cancer. Aurora, CO: International Association for the Study of Lung Cancer; 2017. [Google Scholar]
- 24.de Ruiter EJ, Mulder FJ, Koomen BM, et al. Comparison of three PD-L1 immunohistochemical assays in head and neck squamous cell carcinoma (HNSCC). Mod Pathol. 2020. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 25.Hendry S, Byrne DJ, Wright GM, et al. Comparison of four PD-L1 immunohistochemical assays in lung cancer. J Thorac Oncol. 2018;13:367–376. [DOI] [PubMed] [Google Scholar]
- 26.Cooper WA, Russell PA, Cherian M, et al. Intra- and interobserver reproducibility assessment of PD-L1 biomarker in non-small cell lung cancer. Clin Cancer Res. 2017;23:4569–4577. [DOI] [PubMed] [Google Scholar]
- 27.Hodgson A, Slodkowska E, Jungbluth A, et al. PD-L1 immunohistochemistry assay concordance in urothelial carcinoma of the bladder and hypopharyngeal squamous cell carcinoma. Am J Surg Pathol. 2018;42:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butter R, t Hart NA, Hooijer GKJ, et al. Multicentre study on the consistency of PD-L1 immunohistochemistry as predictive test for immunotherapy in non-small cell lung cancer. J Clin Pathol. 2020;73:423–430. [DOI] [PubMed] [Google Scholar]
- 29.Dobbin KK, Cesano A, Alvarez J, et al. Validation of biomarkers to predict response to immunotherapy in cancer: volume II—clinical validation and regulatory considerations. J Immunother Cancer. 2016;4:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnitt SJ, Connolly JL, Tavassoli FA, et al. Interobserver reproducibility in the diagnosis of ductal proliferative breast lesions using standardized criteria. Am J Surg Pathol. 1992;16:1133–1143. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 Immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017;12:208–222. [DOI] [PubMed] [Google Scholar]
- 32.Torlakovic E, Lim HJ, Adam J, et al. “Interchangeability” of PD-L1 immunohistochemistry assays: a meta-analysis of diagnostic accuracy. Mod Pathol. 2020;33:4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parra ER, Villalobos P, Mino B, et al. Comparison of different antibody clones for immunohistochemistry detection of programmed cell death ligand 1 (PD-L1) on non-small cell lung carcinoma. Appl Immunohistochem Mol Morphol. 2018;26:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodson A, Parry S, Lissenberg-Witte B, et al. External quality assessment demonstrates that PD-L1 22C3 and SP263 assays are systematically different. J Pathol Clin Res. 2020;6:138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzgibbons PL, Bradley LA, Fatheree LA, et al. Principles of analytic validation of immunohistochemical assays: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2014;138:1432–1443. [DOI] [PubMed] [Google Scholar]
- 36.Lawson NL, Dix CI, Scorer PW, et al. Mapping the binding sites of antibodies utilized in programmed cell death ligand-1 predictive immunohistochemical assays for use with immuno-oncology therapies. Mod Pathol. 2020;33:518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Pathology Accreditation Advisory Council (NPAAC). The Requirements for Laboratories Reporting Tests for the National Cervical Screening Program, 2nd ed. Canberra, Australia: Australian Government Department of Health; 2019. [Google Scholar]
- 38.Conant JL, Gibson PC, Bunn J, et al. Transition to subspecialty sign-out at an academic institution and its advantages. Acad Pathol. 2017;4:2374289517714767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jakate K, De Brot M, Goldberg F, et al. Papillary lesions of the breast: impact of breast pathology subspecialization on core biopsy and excision diagnoses. Am J Surg Pathol. 2012;36:544–551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.ajsp.com.


