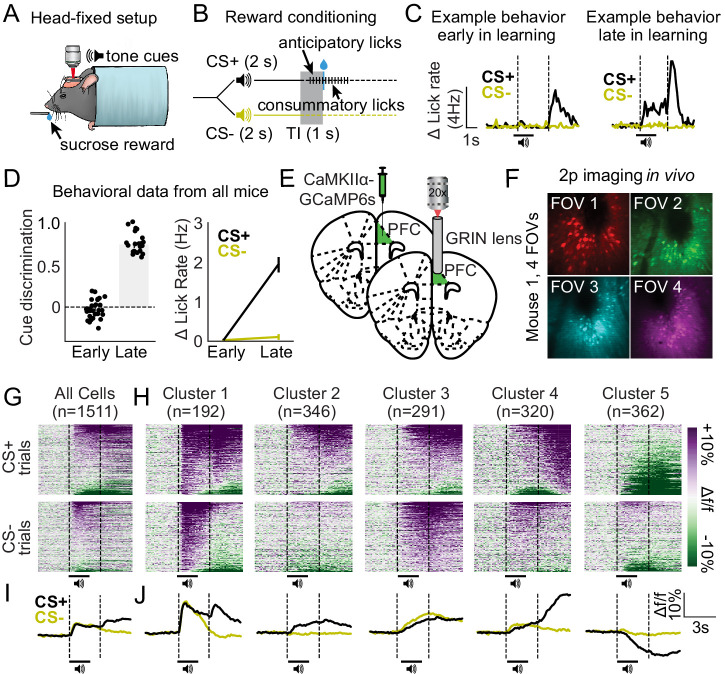
Figure 1. Distinct excitatory neuronal ensembles revealed in dorsomedial prefrontal cortex (dmPFC) during head-fixed Pavlovian conditioning.
(A) Illustration of head fixation for reward conditioning experiments with concurrent two-photon imaging. (B) Schematic for reward conditioning experiments, in which the CS+ and CS- are presented in a random order 50 times each. The CS+ denotes the availability of a liquid sucrose reward following a 1 s trace interval (TI). Anticipatory licks are seen during the trace interval in well-trained mice. (C) Example behavior data during early in learning (left) vs. late in learning (right) behavioral sessions. (D) Cue discrimination scores (auROC; CS+ vs. CS-) and change in lick rate for each cue during early and late in learning behavioral sessions. Data presented as mean ± SEM. (E–F) Surgery schematic (E) allowing for in vivo imaging of GCaMP6s-expressing neurons (F). (G–H) Heat maps displaying the responses of all dmPFC neurons (G) and responses separated by cluster (H) aligned to the cues. (I–J) Line plots displaying the mean activity traces of all cells (I) and mean activity of all cells separated by cluster (J).
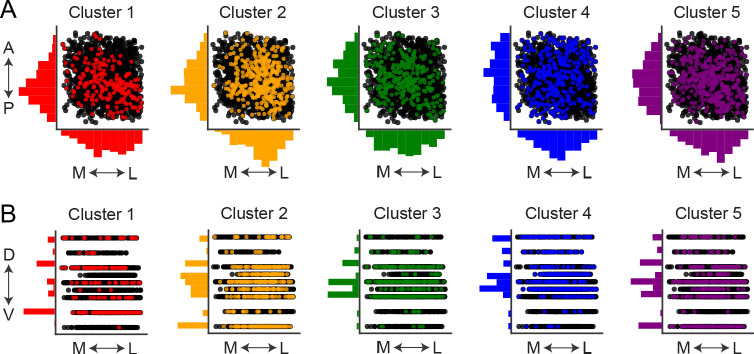
Figure 1—figure supplement 1. Behavioral task acquisition across days for each mouse.
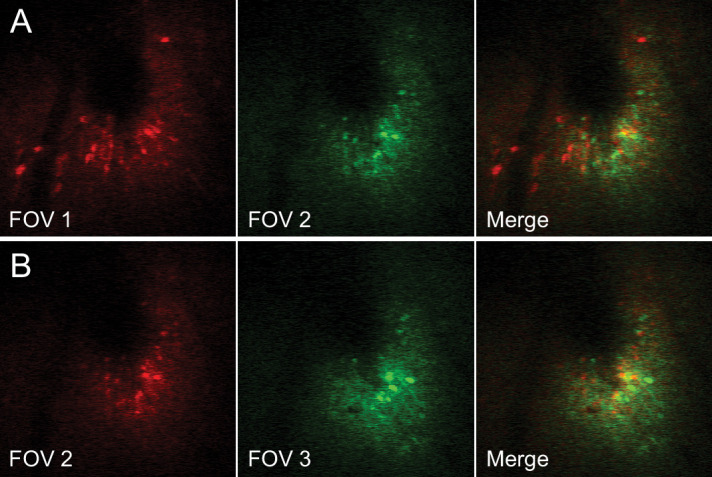
Figure 1—figure supplement 2. Visualizing dorsomedial prefrontal cortex (dmPFC) excitatory neurons within unique fields of view (FOVs) in a single mouse.