Figure 1.
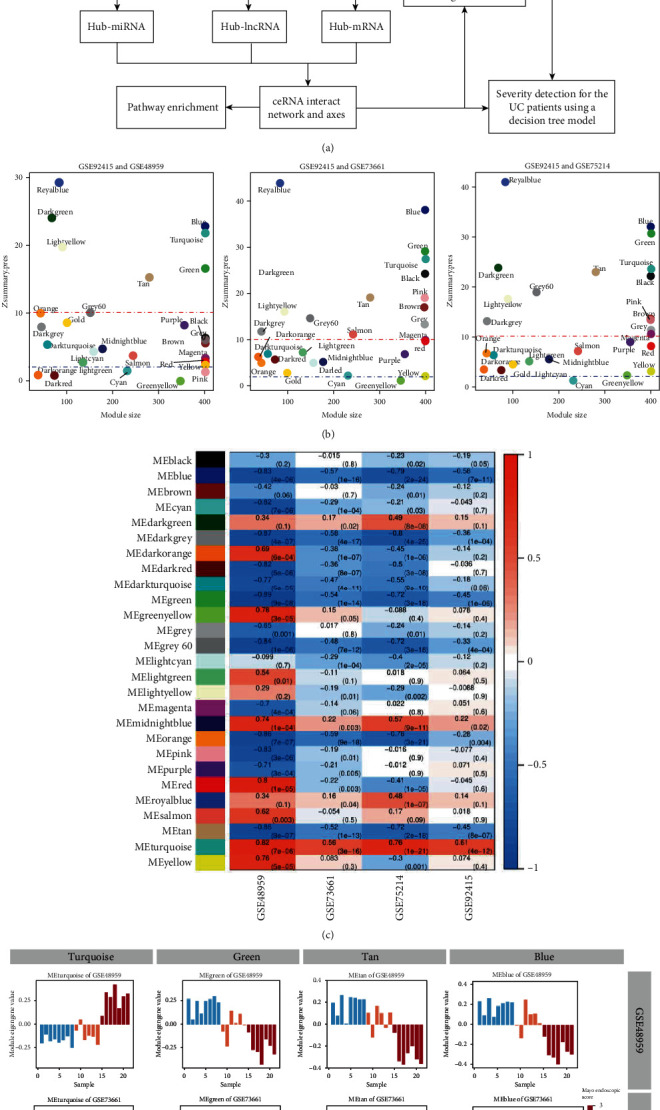
Preprocessing of the lncRNA/mRNA datasets and construction of weighted gene coexpression networks. (a) Flowchart for an overview of the present analysis. (b) Preservation of GSE92415 network modules in different datasets. The y-axis displays the Z score for each module. Labels beside each module (colored dot) represent the corresponding module in the reference dataset. The x-axis represents the number of genes in the module. Z scores of less than 2 (blue bottom line) imply no evidence for module preservation, while scores exceeding 10 (red line) imply strong evidence for module preservation. (c) Module trait relationship: a matrix with the module-trait relationships (MTRs) (correlation coefficients) and corresponding P values (in brackets) between modules on the y-axis and disease progression traits of four datasets on the x-axis. (d) The relationship between the module eigengene and disease severity: the y-axis represents the module eigengene expression value of the sample, and the samples are sorted from mild to severe disease status.
