Figure 1. Schematic Models for Scap and Insig.
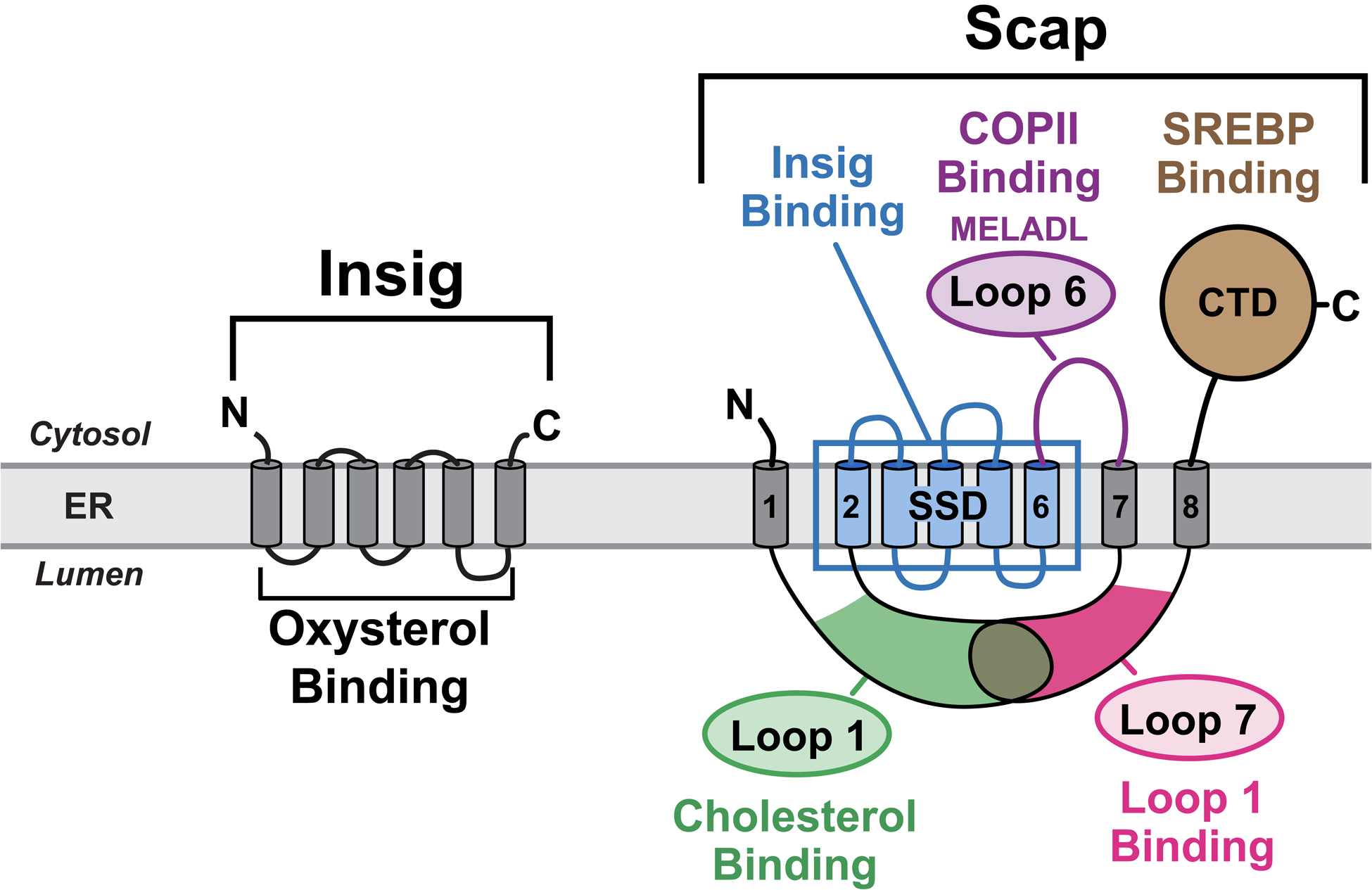
Scap is an ER cholesterol-sensing transmembrane protein with 5 functional subdomains: i) Cholesterol-binding domain (green) localized to luminal Loop 1; ii) Insig-binding domain (blue) localized to TM helices 2–6, which shares homology with other cholesterol regulatory proteins and has thus been defined as the sterol-sensing domain (SSD); iii) COPII-binding domain (purple) localized to cytoplasmic Loop 6 that contains the hexapeptide MELADL, which is recognized by Sec24 proteins for COPII transport to Golgi; iv) Loop1-binding domain localized to luminal Loop 7; and v) SREBP-binding domain localized to the cytoplasmic C-terminal domain (CTD), which contains WD-40 repeats that mediate binding to SREBPs. Insig is an ER oxysterol-sensing transmembrane protein containing six TM helices. Together, Scap and Insig respond to increases in either exogenously-derived cholesterol or endogenously-synthesized oxysterol by forming a Scap/Insig complex, which prevents binding to COPII proteins, blocking the transport of Scap and SREBPs to the Golgi, and reducing SREBP-induced lipid synthesis.
See also Figure S1.
