Figure 5. Structural Basis for the Scap-Insig Interaction.
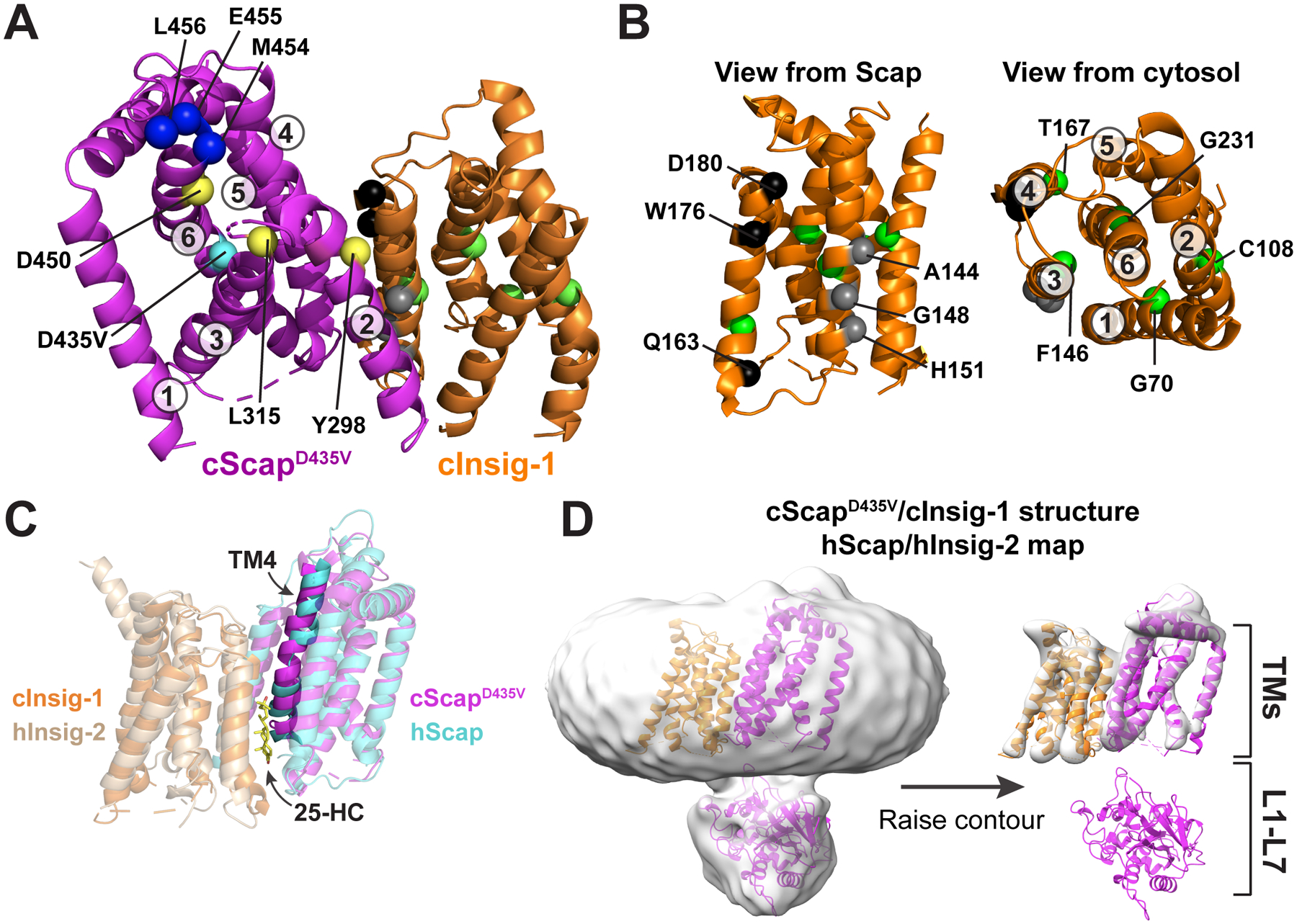
(A) Model of the TM domains of cScapD435V and cInsig-1. cScapD435V is colored magenta and cInsig-1 is colored orange. The Cα atoms of key residues are depicted as spheres. cScap’s TM helices are numbered and the position of key residues are indicated. M454, E455, and L456, shown as blue spheres, are the first visible residues in loop 6 and represent the first 3 of the 6 amino acids of the MELADL motif that is required for COPII binding. Y298, L315, and D450, shown as yellow spheres, render Scap incapable of binding Insig-1 when mutated to the residues indicated in Table S2. The position of the D435V mutation, shown as a cyan sphere, traps Scap in the Insig-binding conformation. See (B) for an explanation of highlighted residues in cInsig-1.
(B): Left: The structure of cInsig-1, viewed from the membrane level as if from cScap. The location of key residues are labeled with black, grey, and green spheres, and are described in Table S3. Black spheres indicate the locations of residues that when mutated as indicated in Table S3 reduce binding to Scap but not to 25-HC. Grey spheres indicate residues that when mutated as indicated in Table S3 lose binding to Scap. Right: cInsig-1 rotated ~90° towards the reader relative to the orientation at bottom left, such that cInsig-1 is viewed from the cytosol. The TM helices are labeled, as are the locations of residues, shown in green spheres, which when mutated as indicated in Table S3 reduce binding to 25-HC.
(C) Comparison of the cScapD435V/cInsig-1 complex with the human Scap/Insig-2/25-HC complex (Yan et al. 2021). Superposition of the transmembrane helices of cScapD435V/cInsig-1 with human Scap/Insig-2/25-HC. Proteins are colored as indicated with transparent cartoons except for TM4. hScap, human Scap; hInsig-2, human Insig-2.
(D) Comparison of the cScapD435V/cInsig-1 structure with the cryo-EM map for the human Scap/Insig-2/25-HC complex. The map (EMD-30074) was gaussian filtered to 3 SD using Chimera, and the cScapD435V/cInsig-1 was docked into the density. The map is contoured at level 0.131 (left) and 0.358 (right). hScap, human Scap; hInsig-2, human Insig-2.
