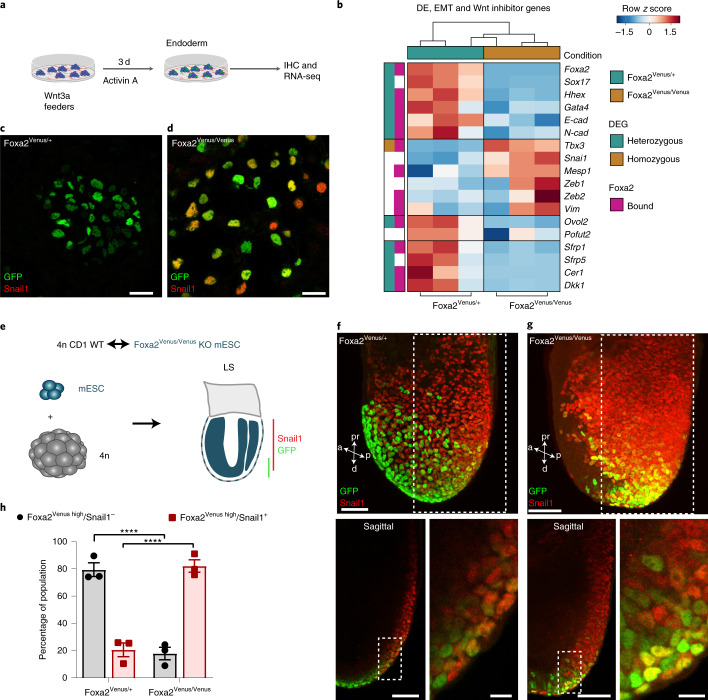
Fig. 4. Foxa2 suppresses Snail1 in the endoderm lineage.
a, Schematic of endoderm differentiation of Foxa2Venus/Venus knockout mESCs. IHC, immunohistochemistry. b, Heatmap of Foxa2Venus/+ and Foxa2Venus/Venus endoderm34 showing the up- and downregulation of endoderm genes, EMT activators, inhibitors and Wnt signalling inhibitors (n = 3 independent differentiations). The pink boxes indicate genes bound by Foxa2. The green and brown boxes mark genes with significantly different expression in Foxa2Venus/+ and Foxa2Venus/Venus cells, respectively. c,d, Immunostainings against Venus and Snail1 of Foxa2Venus/+ control (c) and Foxa2Venus/Venus knockout mESCs (d) differentiated under endoderm conditions for 3 d. e, Schematic of aggregation chimeras generated with Foxa2Venus/+ and Foxa2Venus/Venus mESCs. LS, late-streak stage; WT, wild type. f,g, Maximum projections (top) and single optical sections (bottom) from confocal images of Foxa2Venus/+ (f) and Foxa2Venus/Venus late-streak stage mESC tetraploid aggregation chimeras (g) stained for Venus and Snail1. h, Quantification of Venushigh and Snail1+ or Snail1– cells in Foxa2Venus/+ and Foxa2Venus/Venus aggregation chimeras (****P < 0.0001; ordinary one-way analysis of variance with Tukey’s multiple comparisons test; n = 3 embryos). Statistically non-significant results are not indicated in the figure. All samples were derived from biologically independent experiments. The images in c and d are representative of four independent endoderm differentiations. The images in f and g are representative of three embryos each. The data are presented as mean values ± s.e.m. Scale bars, 50 µm (insets, 10 µm).