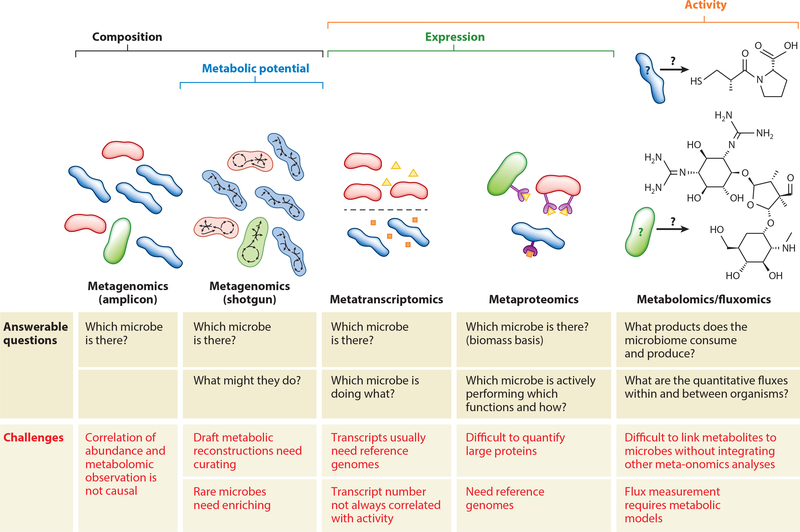
Figure 2.
Different meta-omics tools are suited to answer different questions about microbiome composition and function. Amplicon metagenomics can reveal which organisms are present in a microbiome but not necessarily what each microbe’s role in the community is. Shotgun metagenomics elucidates which microbes are present in the community and what functions they have the capacity to perform. Metatranscriptomics and metaproteomics are necessary to uncover which functions are actually being performed in the community; assigning these transcripts and proteins to the microbes that produced them typically requires high-quality reference genomes or concurrent metagenomics analyses. Metabolomics and fluxomics quantify the chemical composition of the microbiome environment; however, linking metabolites to the microbes that produce or consume them is challenging, even with reference genomes. Linking microbiome composition and function is facilitated by integrating multiple meta-omics techniques, for example, concurrent shotgun metagenomics, metaproteomics, and metabolomics studies to assess which enzymes are producing an observed small molecule of interest and which microbes could produce those enzymes.