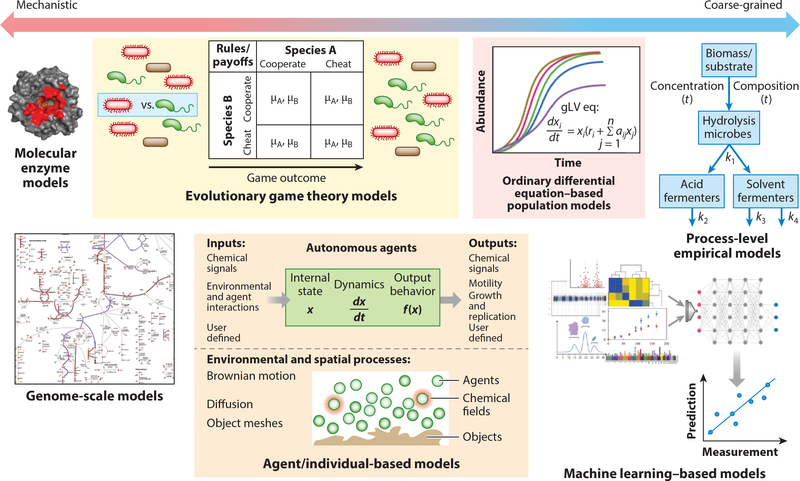
Figure 3.
Microbiomes can be modeled on many scales, and the choice of modeling technique depends on the question at hand. At the most mechanistic level, molecular simulations may be used to model the thermodynamics and kinetics of individual enzymes identified through metaproteomics; however, these are not scalable to encompass the entire microbiome. GEMs enable prediction of the metabolic fluxes and end-product profiles within a microbiome and can offer mechanistic insight into metabolomic observations given high-quality genomic reconstructions and sufficient experimental model validation. Evolutionary game theory models and differential equation–based models are particularly useful when microbiome population dynamics are of the greatest interest, because detailed metabolic reconstructions are not needed for each organism to be modeled. AbMs offer flexibility in that the user may define which inputs and outputs to include in the model and are often the technique of choice when integrating both metabolic and physical interactions between microbes. Data-driven models, including emerging machine learning–based models, offer empirical predictions of microbiome behaviors under specified conditions given appropriate training data. Although less mechanistic than GEMs, machine learning–based models are a pragmatic approach to synthesizing large amounts of different data types into interpretable conclusions, for example, rate constant estimations for process-level models of microbiome function. The structure of the molecular enzyme model is from PDB ID 4QLK. The structure of the AbM is reproduced with permission from Reference 162. The microbes in the evolutionary game theory models panel and the entire machine learning–based models panel were adapted from images created with BioRender.com. Abbreviations: AbM, agent-based model; EGT, evolutionary game theory; GEM, genome-scale model; ODE, ordinary differential equation; PDB, Protein Data Bank.