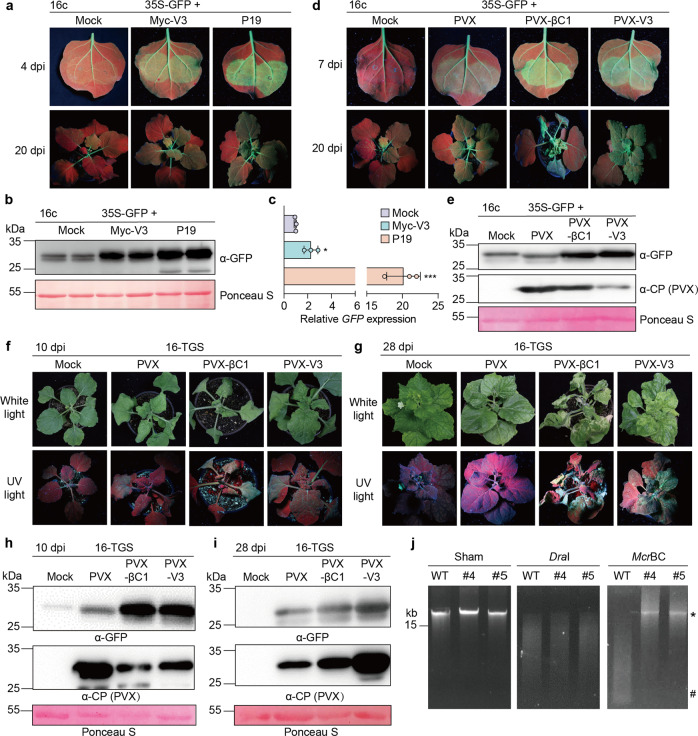
Fig. 5. V3 functions as a suppressor of PTGS and TGS.
a Transgenic 16c N. benthamiana plants co-infiltrated with constructs to express GFP (35S-GFP) and Myc-V3, P19 (as positive control), or mock (empty vector, as negative control) at 4 dpi (upper panel) or 20 dpi (lower panel) under UV light. b Western blot showing the GFP accumulation in inoculated leaves from (a) at 4 dpi. The corresponding Ponceau S staining of the large RuBisCO subunit serves as a loading control. This experiment was repeated three times with similar results; representative images are shown. c Relative GFP mRNA accumulation in inoculated leaves from (a) at 4 dpi measured by qRT-PCR. Data are the mean of three independent biological replicates. Error bars represent SD. An asterisk indicates a statistically significant difference according to unpaired Student’s t test (two-tailed), * p < 0.5, *** p < 0.001. NbActin2 was used as the internal reference. d Transgenic 16c N. benthamiana plants co-infiltrated with constructs to express GFP (35S-GFP) and PVX, PVX-V3, PVX-βC1 (as positive control), or mock (infiltration buffer, as negative control) at 7 dpi (upper panel) or 20 dpi (lower panel) under UV light. e Western blot showing the GFP accumulation in inoculated leaves from (d) at 7 dpi. The corresponding Ponceau S staining of the large RuBisCO subunit serves as loading control. This experiment was repeated three times with similar results; representative images are shown. f, g Symptoms of 16-TGS N. benthamiana plants infected with PVX, PVX-V3, PVX-βC1 (as positive control), or mock-inoculated under white light or UV light at 10 dpi (f) and 28 dpi (g). h, i Western blot showing accumulation of GFP and PVX CP in systemically infected leaves from (f) and (i). The corresponding Ponceau S staining of the large RuBisCO subunit serves as loading control. This experiment was repeated three times with similar results; representative images are shown. j DNA methylation analysis by restriction enzyme digestion in V3-YFP transgenic N. benthamiana plants. Genomic DNA extracted from WT N. benthamiana or two independent V3-YFP transgenic lines (#4 and #5) was digested with the methylation-dependent restriction enzyme McrBC and the methylation-insensitive enzyme DraI. “Sham” indicates a mock digestion with no enzyme added. The positions of undigested input genomic DNA is indicated with an asterisk; the position of the McrBC-digested products is indicated with a hashtag. This experiment was repeated three times with similar results; representative images are shown. The original data from all experiments and replicates can be found in the Source data file.