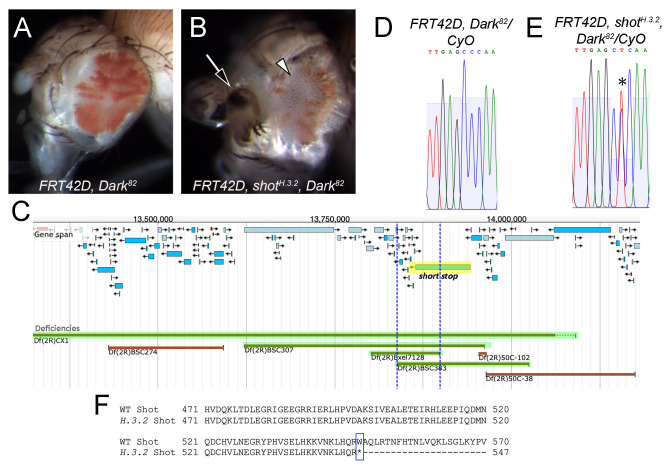
Figure 1. Characterization of shotH.3.2 mutation by phenotypic analysis, complementation mapping, and genetic sequencing.
FRT42D, Dark82 control mosaic eyes (A) exhibit an increased ratio of red (mutant) to white (wildtype) tissue compared to FRT42D, shotH.3.2, Dark82mosaic eyes (B). Mosaic eyes with shotH.3.2, Dark82 clones display antennal defects (arrow) and disorganized wildtype ommatidia (arrowhead). (C) Map of deficiency lines that complemented (red bars) and failed to complement (highlighted in green) the H.3.2 mutation, resulting in a region of overlap of 2R:13,839,479..13,897,827 (between blue dashed lines). short stop (shot) is highlighted in yellow. Adapted from FlyBase using release FB2021_03 (Larkin et al. 2021). (D-E) The nucleotide location of H.3.2 was identified by sequence analysis. FRT42D, Dark82 control sequence with a single C peak at 2R:13,904,428 in the coding region of shot (D), compared to FRT42D, shotH.3.2, Dark82 mutant sequence with a heterozygous double peak revealing a nucleotide change from C to T at this position (E, asterisk). (F) Amino acid alignment of wildtype (WT) and H.3.2 mutant Shot sequences. Amino acids 471 to 570 encode a spectrin-like repeat in Shot isoform H. The Trp548* mutation in the H.3.2 mutant allele is indicated (box).