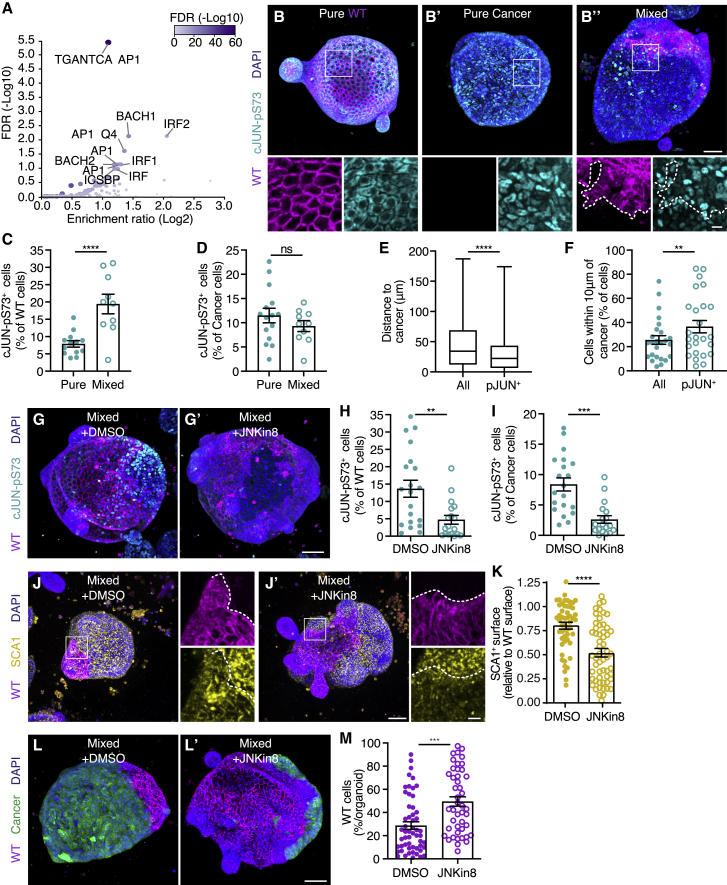
Figure 6.
JNK signaling drives cell competition
(A) Transcription factor target analysis of differentially expressed genes (p < 0.05) that are enriched in mixed wild-type cells. The graph displays enrichment (Log2) and false discovery rate (FDR) (−Log10) of gene sets (significantly enriched gene sets are indicated).
(B) Representative 3D-reconstructed confocal images of pure WT (B), pure cancer (B’), and mixed (B’’) organoids, stained for activated cJUN (cyan); nuclei are visualized with DAPI (blue). The insets display a 3× magnification of the area in the white box.
(C and D) Quantification of the number of cJUN-pS73+ cells relative to the total number of wild-type (C) and cancer (D) cells; each dot represents one organoid (mean ± SEM; one-way ANOVA; multiple comparisons; p < 0.0001, n = 13 and 10 organoids, C; p = 0.5740, n = 15 and 10 organoids, D).
(E) Quantification of the distance of all (left) and cJUN-pS73+ (right) wild-type nuclei to the closest cancer cell surface in μm (median; 25th to 75th percentiles [box]; min. to max. [whiskers]; unpaired t test; two-tailed; p < 0.0001; n = 25 organoids).
(F) Quantification of wild-type cells within a one-cell diameter (10 μm) of the closest cancer cell surface. The percentages of all (left) and cJUN-pS73+ (right) wild-type cells at the interface are displayed (mean ± SEM; paired t test; two-tailed; p = 0.0093; n = 25 organoids).
(G–I) Representative 3D-reconstructed confocal images of control (G) and JNKin8 treated (G’) mixed organoids stained for activated cJUN (cyan); nuclei are visualized with DAPI (blue).
(H and I) Quantification of the number of cJUN-pS73+ cells relative to the total number of wild-type (H) and cancer (I) cells; each dot represents one organoid (mean ± SEM; one-way ANOVA; multiple comparisons; p = 0.0041, n = 20 and 18 organoids, H; p = 0.0001, n = 20 and 18 organoids, I).
(J and K) Representative 3D-reconstructed confocal images of control (J) and JNKin8 treated (J’) mixed organoids and quantification of the SCA1+ surface relative to the total wild-type surface area. The organoids were stained for SCA1 (yellow); nuclei are visualized with DAPI (blue). The insets display a 3.5× magnification of the area in the white box. Each dot in (K) represents one organoid (mean ± SEM; non-parametric; ANOVA; multiple comparisons: p < 0.0001; n = 52 and 56 organoids).
(L and M) Representative 3D-reconstructed confocal image of control (L) and JNKin8 treated (L’) mixed organoids, nuclei are stained with DAPI (blue), and quantification of the percentage of wild-type cells contributing to mixed organoids (M); each dot represents one organoid (mean ± SEM; unpaired t test; two-tailed; p < 0.0001; n = 55 and 45 organoids).
Scale bars represent 50 μm, excluding magnifications in (B) and (J), where scale bar represents 10 μm. See also Figure S6.