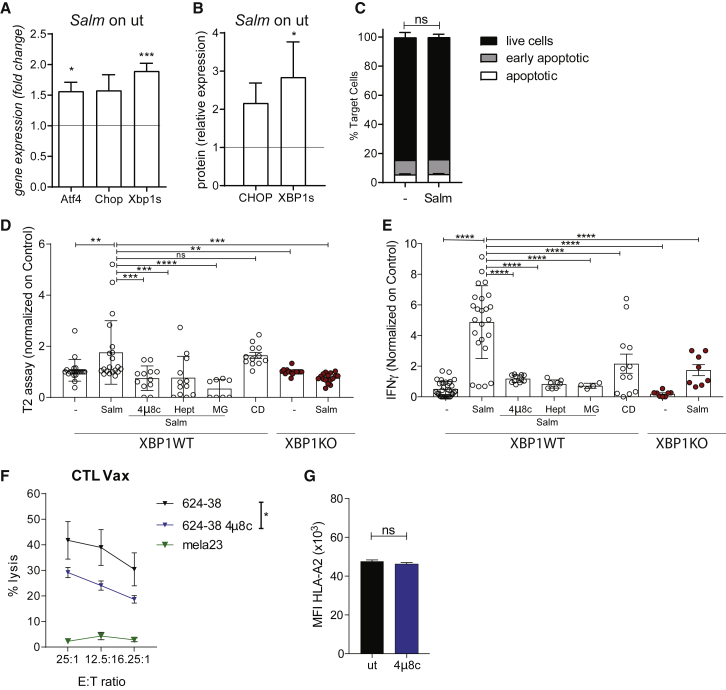
Figure 4.
Salmonella-induced secretome peptides are mainly tumor proteasome derived and are released in response to UPR pathway activation along with hemichannel opening
(A and B) UPR pathway analysis at gene (A) and protein (B) level in Salmonella-infected 624-38 cells. Data of 3 pooled experiments are normalized to Gapdh (A) or Vinculin (B) and expressed as mean ± SEM of average expression fold change.
(C) Frequency of Annexin−PI− (live), Annexin+PI− (early apoptotic), and Annexin+PI+ (apoptotic) tumor cells Salmonella-infected (Salm) or untreated (−) (n = 2).
(D) MFI of HLA-A∗02:01 on T2 cells loaded with secretomes from untreated (−) or Salmonella-infected (Salm) XBP1WT or XBP1KO cells; from Salmonella-infected XBP1WT in combination with 4μ8c, Heptanol (Hept), MG132 (MG); supernatant of dead cells (C and D).
(E) ELISA of IFN-γ released by CTL-Vax stimulated with secretomes differently derived. (D and E) Data of 3 pooled experiments normalized on (−) are represented as mean ± SEM using a scatter dot plot.
(F) Delfia assay; lysis of target cells by CTL-Vax (effector) at different effector-to-target (E:T) ratios (n = 3).
(G) MFI of HLA-A∗02:01 on 624-38 cells. Data of 3 pooled experiments are represented.
Statistical analysis was evaluated using two-sided Mann-Whitney test (A, B, C, and G), one-way ANOVA followed by multiple comparisons (Dunnett) against XBP1WT-Salm (D and E), or two-way ANOVA followed by multiple comparisons (Tukey) (F). ∗p < 0.05,∗∗p < 0.01,∗∗∗p < 0.001,∗∗∗∗p < 0.0001. See also Figures S4, S5, and S6.