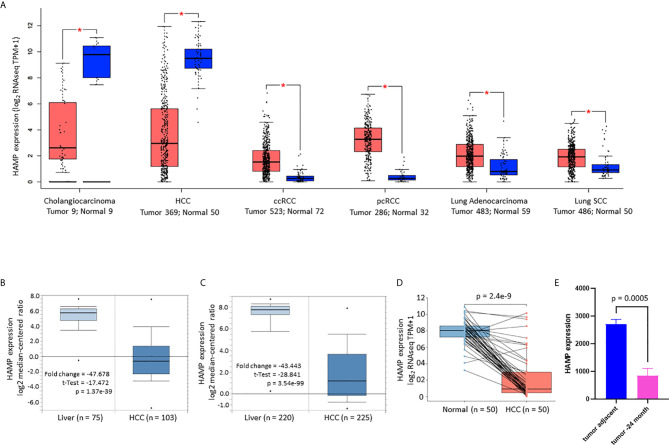
Figure 1.
Hepcidin expression is drastically reduced in liver cancers. (A) Hepcidin expression profiles in human cancers were compared with normal tissues using the TCGA dataset on the GEPIA platform (19). Case numbers in each group were listed at the bottom of the figure. The asterisks indicate a statistically significant difference (p < 0.05, Wilcoxon rank-sum test). HCC, hepatocellular carcinoma; ccRCC, clear cell renal cell carcinoma; pcRCC, papillary cell renal cell carcinoma; Lung SCC, lung squamous cell carcinoma. (B, C) Hepcidin expression in liver cancers was assessed in two cDNA microarray datasets (37, 38). The comparison between normal and malignant liver tissues was conducted using a Student t-test. (D) Hepcidin expression in human liver cancers was compared with matched tumor-adjacent benign tissues in 50 cases from the TCGA project. The p-value was from a paired t-test. Note: only three cases showed a slight increase in cancer tissue compared to the benign compartment. (E) Hepcidin expression data was derived from the GDS2006 dataset in the Tnixp-deficient mice with spontaneous liver cancer (30). Hepcidin expression levels were normalized with ACTB gene levels as an internal control (n = 3).