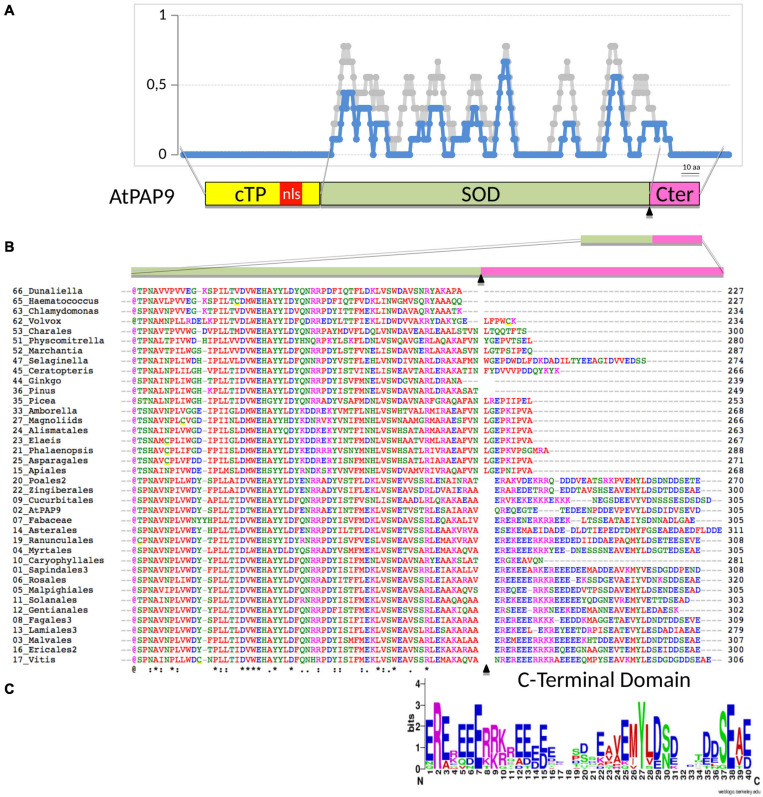
FIGURE 2.
Sequence alignment of predicted orthologous PAP9 protein found in representatives of phylogenetic clades as described in Supplementary Table 1. (A) Amino acid identity (in blue) and similarity (in light gray) given in a 10-aa moving ratio. The alignment corresponds to the full dataset given in Supplementary Table 2 marked with a cross (column named in this figure). Schematic illustration of the PAP9 domains: cTP, chloroplast transit peptide in yellow; NLS, predicted nuclear localization signal in red; SOD, superoxide dismutase domain in khaki; C-terminal domain in magenta. (B) Amino acid partial alignment of the accessions as above, the 2-digit number corresponds to the clade given in Supplementary Table 1. cTP, transit peptide as predicted with ChloroP1.1 (www.cbs.dtu.dk) underlined in yellow and described in Supplementary Table 3. (*), (:), or (.), conserved, strongly similar or weakly similar amino acid properties (standards from www.uniprot.org). Amino acids colors as in Clustal Omega [red (AVFPMILW): small + hydrophobic (includes aromatic Y); blue (DE): acidic; magenta (RHK): basic; green (STYHCNGQ): hydroxyl + sulfhydryl + amine + G). bNLS, bipartite NLS as predicted with NLS mapper (http://nls-mapper.iab.keio.ac.jp). (C) AA sequence logo (Crooks et al., 2004) generated on the C-terminal domain alignment of PAP9 with the 40-aa-long stretch from clade 22 (Zingiberales) to clade 17 (Vitis) excluding clade 10 (Caryophyllales).