FIG. 3.
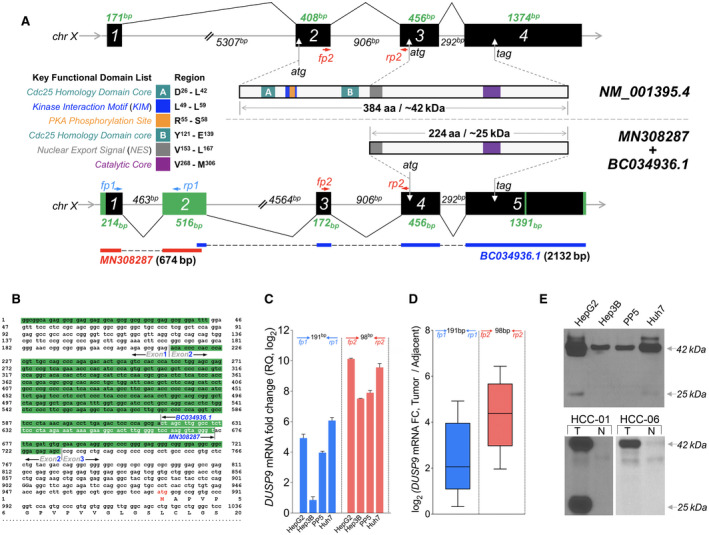
Identification of HCC‐specific DUSP9 isoform. (A) Schematic illustration comparing WT DUSP9 (NM_001395.4 at top) and N‐terminal truncated DUSP9 isoform (bottom, combination of MN308287 and BC034936.1). Black/green boxes indicate exons. Coding sequences are shown with key functional domains. Green segments in truncated isoform represent sequence differences compared to WT DUSP9. Primer pairs (fp1/rp1, fp2/rp2) are also indicated. (B) Combination of 5’RACE‐PCR amplified fragment (MN) with published partial sequence BC034936.1. MN308287 consists of g1 to t674, including 57 nucleotides of overlap with BC034936.1 (white letters), revealing 617 nucleotides of novel 5’ sequence. Red “atg” represents the predicted translation initiation site of the HCC‐specific DUSP9 isoform. (C) qPCR using primers selective for the truncated DUSP9 isoform (fp1/rp1, blue bars) or all DUSP9 isoforms (fp2/rp2, red bars) in HCC cell lines (RQ normalized to DUSP9 mRNA level in THLE‐2). Graph shows mean + SEM. (D) qPCR using primers selective for the truncated DUSP9 isoform (fp1/rp1, blue box) or all DUSP9 isoforms (fp2/rp2, red box) in 10 HCC tumors and adjacent liver tissues. Graph shows IQR (box), median (horizontal line), and outliers (whiskers). (E) Western blot of DUSP9 in four HCC cell lines, human HCC tumors, and adjacent liver. Abbreviations: aa, amino acid; Cdc25, cell division cycle 25; chr, chromosome; FC, fold change; N, adjacent liver; RQ, relative quantification; T, HCC tumors.
